Eelco Tromer
@eelcotromer.bsky.social
NWO-Veni Fellow @GBB Rijksuniversiteit van Groningen | eukaryotic diversity | chromosome segregation | evolutionary cell biology | exploring meiosis
New work on ARK1 (AURKB?) in Plasmodium (malaria parasite) with @ritatewari.bsky.social and Pushkar Sharma. CPC in Plasmodium is (ofcourse..) different compared to conventional models: two INCENPs, no borealin/survivin - mainly at spindle microtubules/MTOCs. Special thanks to @ryanase.bsky.social!
Pleased to share our new preprint: "Plasmodium ARK1 regulates spindle formation during atypical mitosis and forms a divergent chromosomal passenger complex". Many thanks to our collaborators.
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
August 31, 2025 at 12:26 PM
New work on ARK1 (AURKB?) in Plasmodium (malaria parasite) with @ritatewari.bsky.social and Pushkar Sharma. CPC in Plasmodium is (ofcourse..) different compared to conventional models: two INCENPs, no borealin/survivin - mainly at spindle microtubules/MTOCs. Special thanks to @ryanase.bsky.social!
Reposted by Eelco Tromer
Pleased to share our new preprint: "Plasmodium ARK1 regulates spindle formation during atypical mitosis and forms a divergent chromosomal passenger complex". Many thanks to our collaborators.
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
August 28, 2025 at 1:12 PM
Pleased to share our new preprint: "Plasmodium ARK1 regulates spindle formation during atypical mitosis and forms a divergent chromosomal passenger complex". Many thanks to our collaborators.
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
@ritatewari.bsky.social @eelcotromer.bsky.social @davidguttery.bsky.social
www.biorxiv.org/content/10.1...
Reposted by Eelco Tromer
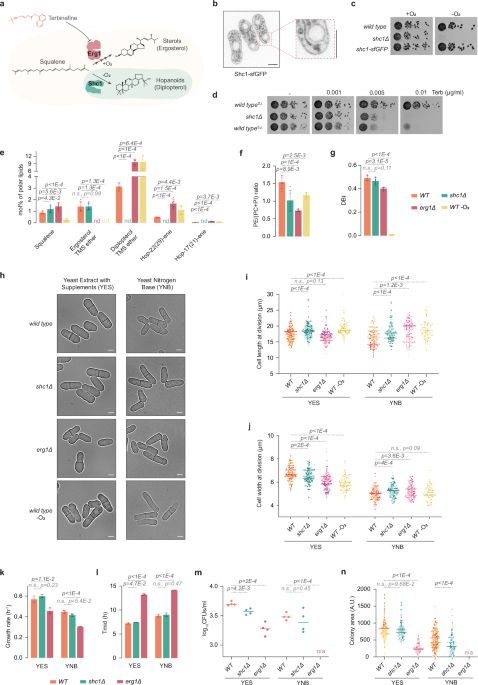
Happy to share the first story from my postdoc in @schnittger-lab.bsky.social
We studied TITAN9, a Csm1-like protein in Arabidopsis.
Even though I now work with soil, chromosome segregation and meiosis still have a special place in my heart.
#meiosis4eva #meiosis #chromosomesegregation
We studied TITAN9, a Csm1-like protein in Arabidopsis.
Even though I now work with soil, chromosome segregation and meiosis still have a special place in my heart.
#meiosis4eva #meiosis #chromosomesegregation
August 3, 2025 at 11:10 AM
Happy to share the first story from my postdoc in @schnittger-lab.bsky.social
We studied TITAN9, a Csm1-like protein in Arabidopsis.
Even though I now work with soil, chromosome segregation and meiosis still have a special place in my heart.
#meiosis4eva #meiosis #chromosomesegregation
We studied TITAN9, a Csm1-like protein in Arabidopsis.
Even though I now work with soil, chromosome segregation and meiosis still have a special place in my heart.
#meiosis4eva #meiosis #chromosomesegregation
Reposted by Eelco Tromer
So excited to share this preprint from the Rosin lab in collaboration with the Hawley lab and @eelcotromer.bsky.social on moth spermatogenesis! We investigate the meiotic errors that occur during the formation of apyrene sperm (that have no DNA) in silkworms!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Programmed meiotic errors facilitate dichotomous sperm production in the silkworm, Bombyx mori
The goal of meiosis is typically to produce haploid gametes (eggs or sperm). Failure to do so is catastrophic for fertility and offspring health. However, Lepidopteran (moths and butterflies) males pr...
www.biorxiv.org
August 4, 2025 at 10:38 AM
So excited to share this preprint from the Rosin lab in collaboration with the Hawley lab and @eelcotromer.bsky.social on moth spermatogenesis! We investigate the meiotic errors that occur during the formation of apyrene sperm (that have no DNA) in silkworms!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Eelco Tromer
Postdoc position currently available in our group studying meiosis in arabidopsis and wheat:
www.jic.ac.uk/vacancies/po...
www.jic.ac.uk/vacancies/po...
July 21, 2025 at 9:02 AM
Postdoc position currently available in our group studying meiosis in arabidopsis and wheat:
www.jic.ac.uk/vacancies/po...
www.jic.ac.uk/vacancies/po...
Reposted by Eelco Tromer
We are growing! A new @zeiss-microscopy.bsky.social widefield tailored for ExM- supported by @moorefound.bsky.social is settling! Another one is on its way at @gautamdey.bsky.social Lab !
These two will become the powerhorses for high-througput imaging of the ExM Atlas of #Microbial Eukaryote !
These two will become the powerhorses for high-througput imaging of the ExM Atlas of #Microbial Eukaryote !

July 17, 2025 at 12:01 PM
We are growing! A new @zeiss-microscopy.bsky.social widefield tailored for ExM- supported by @moorefound.bsky.social is settling! Another one is on its way at @gautamdey.bsky.social Lab !
These two will become the powerhorses for high-througput imaging of the ExM Atlas of #Microbial Eukaryote !
These two will become the powerhorses for high-througput imaging of the ExM Atlas of #Microbial Eukaryote !
Happy to see our work published and glad to have contributed together with @maxraas.bsky.social ! Looking forward to all the projects that will come out of this work!
Excited to share our new paper in @cellreports.bsky.social that reshapes our understanding of chromosome organization's deep evolutionary roots! Our work dives into the origins of the machinery that structures our very genomes.
🔗: doi.org/10.1016/j.ce...
#Genomics #Evolution #CellBiology #LECA
🔗: doi.org/10.1016/j.ce...
#Genomics #Evolution #CellBiology #LECA
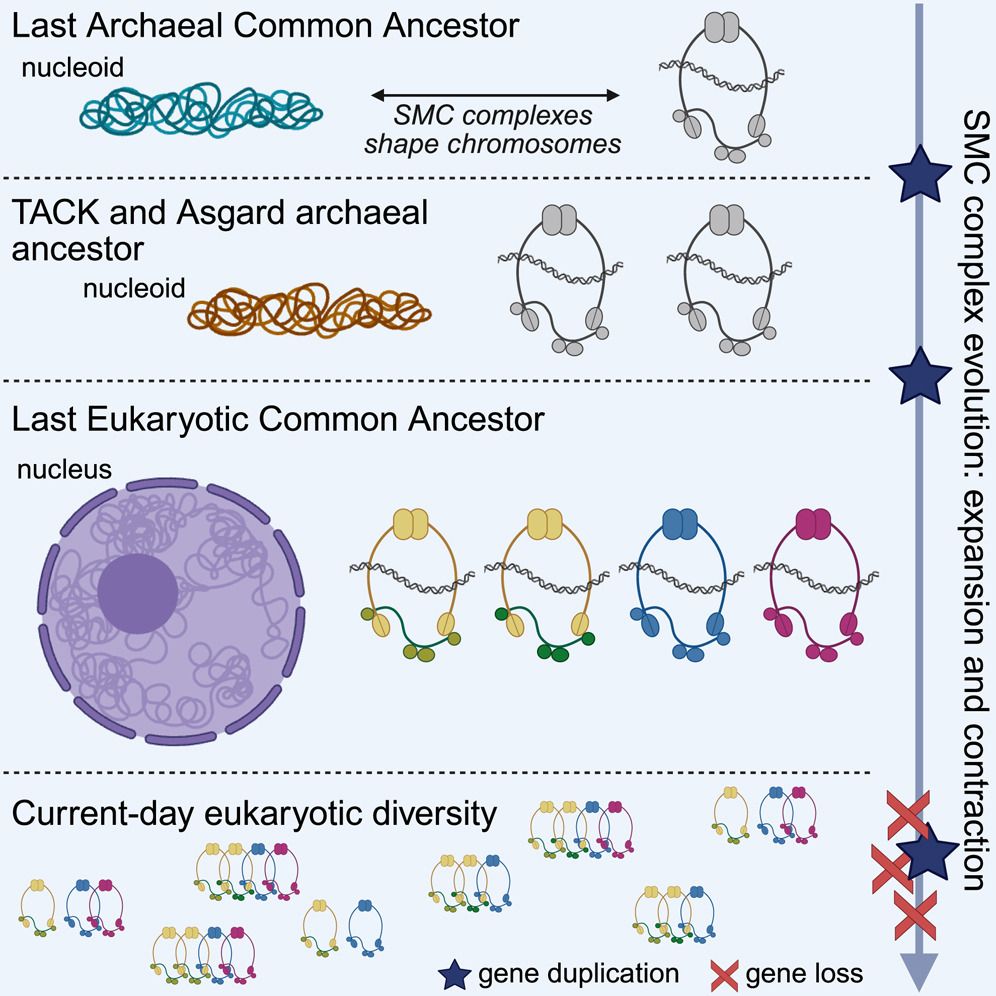
June 23, 2025 at 11:18 AM
Happy to see our work published and glad to have contributed together with @maxraas.bsky.social ! Looking forward to all the projects that will come out of this work!
Reposted by Eelco Tromer
It’s out! The evolutionary origins of yeast point centromeres uncovered!
“Ancient co-option of LTR retrotransposons as yeast centromeres”
www.biorxiv.org/content/10.1...
“Ancient co-option of LTR retrotransposons as yeast centromeres”
www.biorxiv.org/content/10.1...

Ancient co-option of LTR retrotransposons as yeast centromeres
The evolutionary origins of the genetic point centromere in the brewer's yeast Saccharomyces cerevisiae, a member of the order Saccharomycetales, are still unknown. Competing hypotheses suggest that t...
www.biorxiv.org
April 25, 2025 at 1:59 PM
It’s out! The evolutionary origins of yeast point centromeres uncovered!
“Ancient co-option of LTR retrotransposons as yeast centromeres”
www.biorxiv.org/content/10.1...
“Ancient co-option of LTR retrotransposons as yeast centromeres”
www.biorxiv.org/content/10.1...
Reposted by Eelco Tromer
Reposted by Eelco Tromer
I am happy to share a preprint on the marine plankton project I started one year ago (thanks to Julius and Drahomira's help)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
March 24, 2025 at 3:22 PM
I am happy to share a preprint on the marine plankton project I started one year ago (thanks to Julius and Drahomira's help)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Eelco Tromer
Asgardarchaea & Alphaproteobacteria: key players for the assembly eukaryotic central carbon metabolism! Our new study reveals their gene contributions to glycolysis & TCA cycle, supporting synthrophic scenarios of eukaryogenesis.
rdcu.be/eb0aK
rdcu.be/eb0aK
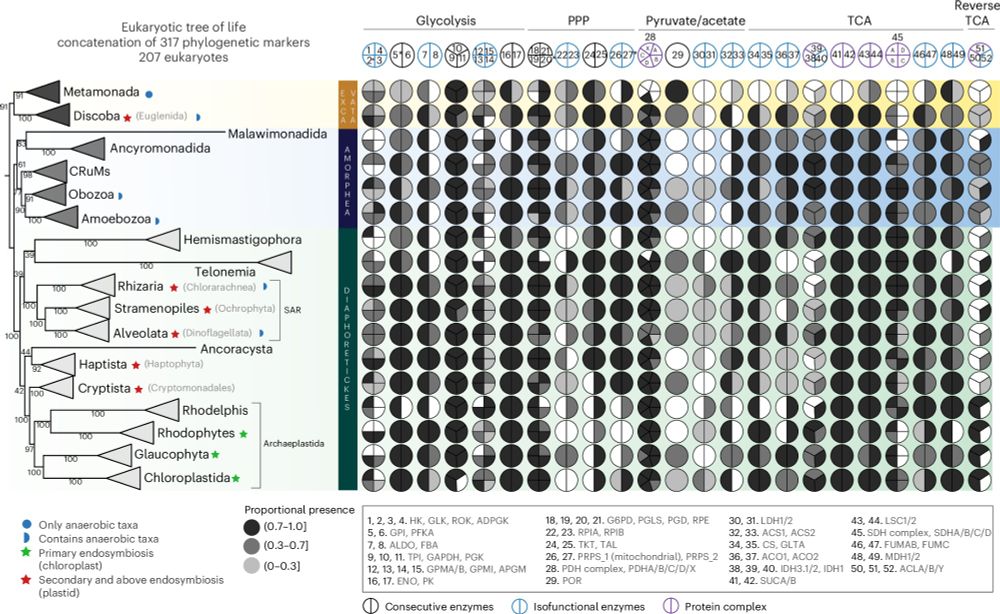
Chimeric origins and dynamic evolution of central carbon metabolism in eukaryotes
Nature Ecology & Evolution - Analysis of the eukaryotic gene repertoires mediating central carbon metabolism identifies ancestral contributions from Alphaproteobacteria, Asgardarchaeota and...
rdcu.be
March 3, 2025 at 1:28 PM
Asgardarchaea & Alphaproteobacteria: key players for the assembly eukaryotic central carbon metabolism! Our new study reveals their gene contributions to glycolysis & TCA cycle, supporting synthrophic scenarios of eukaryogenesis.
rdcu.be/eb0aK
rdcu.be/eb0aK
Reposted by Eelco Tromer
Exciting news! I'm looking for a computational biologist to join my lab at Wageningen University & Research! It's a 3-year postdoc position on Holomycota genome architecture and evolution. Spread the word! www.wur.nl/en/vacancy/p...

Postdoc Comparative genomics and genome architecture in early-diverging fungi and their protist relatives (3-year position)
www.wur.nl
March 5, 2025 at 11:56 AM
Exciting news! I'm looking for a computational biologist to join my lab at Wageningen University & Research! It's a 3-year postdoc position on Holomycota genome architecture and evolution. Spread the word! www.wur.nl/en/vacancy/p...
Reposted by Eelco Tromer
Have questions about #bioinformatics, #genomics, #evolution or stuck on a project? 🧬💻
I want to help!
Happy to discuss data, figures, writing, academia/industry or chatting just for kicks—no strings attached
Drop questions below, DM, or schedule a meeting
Science together is better than alone 🙌
I want to help!
Happy to discuss data, figures, writing, academia/industry or chatting just for kicks—no strings attached
Drop questions below, DM, or schedule a meeting
Science together is better than alone 🙌
December 8, 2024 at 5:53 PM
Have questions about #bioinformatics, #genomics, #evolution or stuck on a project? 🧬💻
I want to help!
Happy to discuss data, figures, writing, academia/industry or chatting just for kicks—no strings attached
Drop questions below, DM, or schedule a meeting
Science together is better than alone 🙌
I want to help!
Happy to discuss data, figures, writing, academia/industry or chatting just for kicks—no strings attached
Drop questions below, DM, or schedule a meeting
Science together is better than alone 🙌
Reposted by Eelco Tromer
Online: our evolutionary interrogation of eukaryotic Structural Maintenance of Chromosomes (SMC) complexes @maxraas.bsky.social @eelcotromer.bsky.social @lauraeme.bsky.social #SMCcomplexes #eukaryogenesis #LECA #condensin #cohesin #evolutionarycellbiology #chromatin 🧵(1/6) doi.org/10.1101/2024...

Shaping up genomes: Prokaryotic roots and eukaryotic diversification of SMC complexes
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
doi.org
January 9, 2024 at 8:36 AM
Online: our evolutionary interrogation of eukaryotic Structural Maintenance of Chromosomes (SMC) complexes @maxraas.bsky.social @eelcotromer.bsky.social @lauraeme.bsky.social #SMCcomplexes #eukaryogenesis #LECA #condensin #cohesin #evolutionarycellbiology #chromatin 🧵(1/6) doi.org/10.1101/2024...
Reposted by Eelco Tromer
A new preprint from our group in collaboration with @jvhooff.bsky.social and @eelcotromer.bsky.social. We discovered that trypanosomes have (a zombie of) Bub1-Bub3 despite the fact that tryps do not have a canonical spindle checkpoint! This is a team work involving Dan, WillC, Midori and Patryk
Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins https://www.biorxiv.org/content/10.1101/2024.01.04.574194v1

Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins https://www.biorxiv.org/content/10.1101/2024.01.04.574194v1
Faithful transmission of genetic material is crucial for the survival of all organisms. In many euka
www.biorxiv.org
January 5, 2024 at 3:43 PM
A new preprint from our group in collaboration with @jvhooff.bsky.social and @eelcotromer.bsky.social. We discovered that trypanosomes have (a zombie of) Bub1-Bub3 despite the fact that tryps do not have a canonical spindle checkpoint! This is a team work involving Dan, WillC, Midori and Patryk