This is where you apply to work with me,
@alexijie.bsky.social @avapamini.bsky.social @lcrawford.bsky.social or Kristen Severson!
(new) link and some instructions below
apply.careers.microsoft.com/careers/job/...

This is where you apply to work with me,
@alexijie.bsky.social @avapamini.bsky.social @lcrawford.bsky.social or Kristen Severson!
(new) link and some instructions below
apply.careers.microsoft.com/careers/job/...

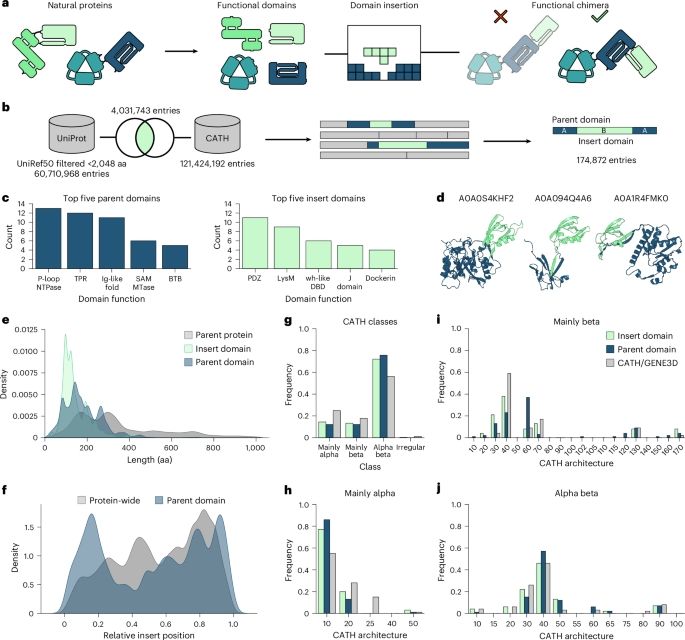
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
doi.org/10.1101/2025...
doi.org/10.1101/2025...
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.

If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.



www.biorxiv.org/content/10.1...
We address a big challenge in synbio: If you give me a protein "X", how can I give you a version of X whose activity is controlled by a kinase?

www.biorxiv.org/content/10.1...
We address a big challenge in synbio: If you give me a protein "X", how can I give you a version of X whose activity is controlled by a kinase?
Front page NYT today, by @kevinroose.com
Gift link www.nytimes.com/2025/01/23/t...

Front page NYT today, by @kevinroose.com
Gift link www.nytimes.com/2025/01/23/t...
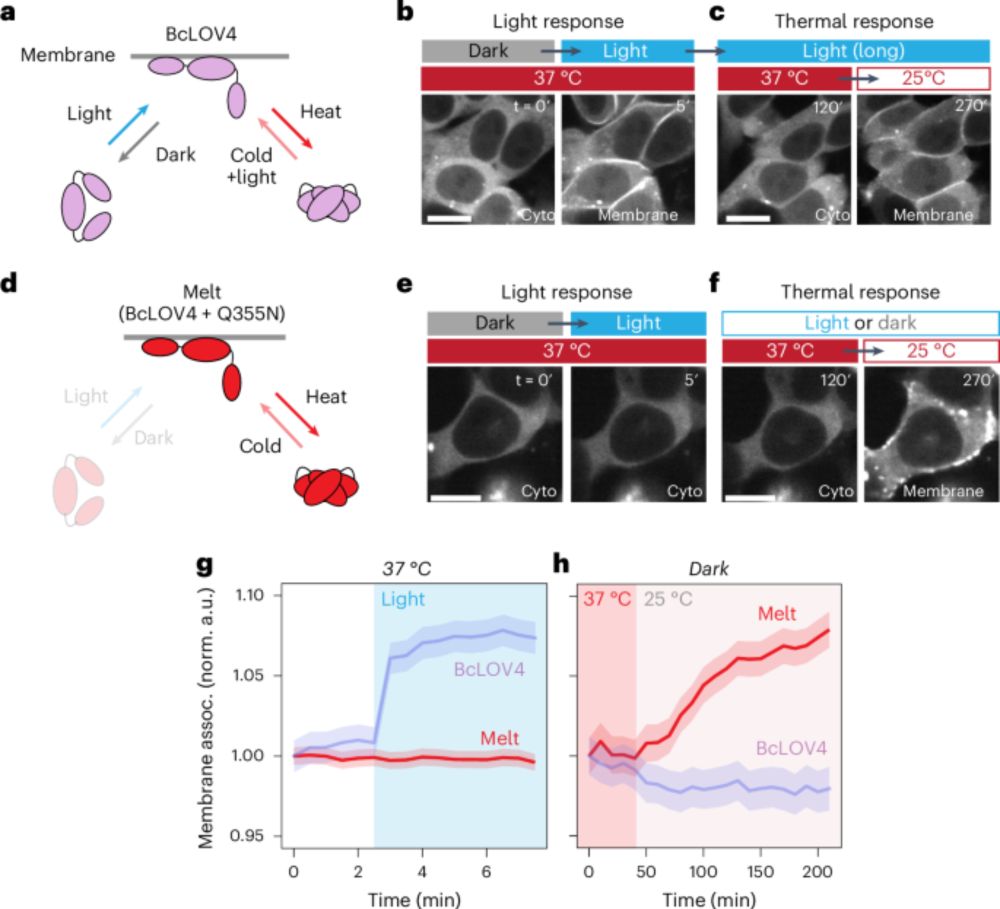
t.co/vtYlhi8aQm
t.co/vtYlhi8aQm

They however fail to naturally sample rare datapoints, like very high activities.
In our new preprint, we show that RL can solve this without the need for additional data:
arxiv.org/abs/2412.12979
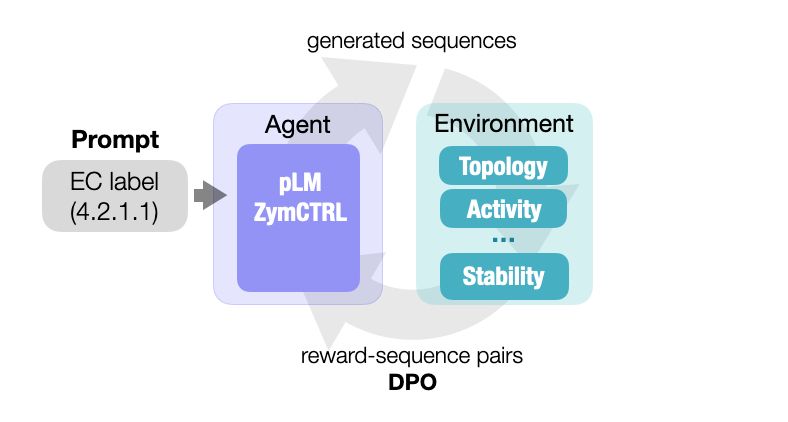
They however fail to naturally sample rare datapoints, like very high activities.
In our new preprint, we show that RL can solve this without the need for additional data:
arxiv.org/abs/2412.12979

Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!

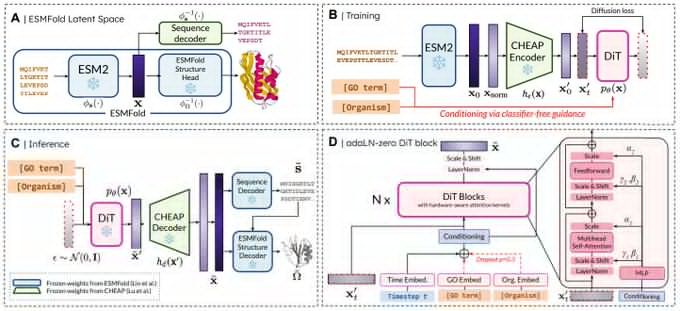

Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!
@schmidts.bsky.social's lab. We use generative #AI
to design new #CARTcell therapies. Check out Andrea's thread & thanks to the whole team! www.biorxiv.org/content/10.1...
@schmidts.bsky.social's lab. We use generative #AI
to design new #CARTcell therapies. Check out Andrea's thread & thanks to the whole team! www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
(1/3)

www.biorxiv.org/content/10.1...
(1/3)
(1/4)

(1/4)


