& how viruses overcome these nucleotide-based immune mechanisms 🗡️
www.nature.com/articles/s41...
The first viral sponges to inhibit Pycsar and type IV Thoeris
Congrats to talented leading author Romi Hadary! Read her thread to learn more about our findings


The first viral sponges to inhibit Pycsar and type IV Thoeris
Congrats to talented leading author Romi Hadary! Read her thread to learn more about our findings
Here we explore how recurrent adaptation of a single anti-defense protein fold enables evasion of distinct immune systems.
Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.

Here we explore how recurrent adaptation of a single anti-defense protein fold enables evasion of distinct immune systems.
Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.

Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.
Paper by @reneechang.bsky.social in @cp-molcell.bsky.social on a nucleotide sponge www.cell.com/molecular-ce...
and preprint by @doudna-lab.bsky.social on viral nucleases www.biorxiv.org/content/10.1...

Paper by @reneechang.bsky.social in @cp-molcell.bsky.social on a nucleotide sponge www.cell.com/molecular-ce...
and preprint by @doudna-lab.bsky.social on viral nucleases www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Together with @reneechang.bsky.social @kranzuschlab.bsky.social and the amazing @soreklab.bsky.social, we explored viral sponges to map their diversity and function.
Discovered huge diversity, including sponges that inhibit Pycsar & Type IV Thoeris!
www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...
Was fun to write this piece with Dina Hochhauser!
Here is a thread to explain the premises
1/
www.nature.com/articles/s41...
Was fun to write this piece with Dina Hochhauser!
Here is a thread to explain the premises
1/
& how viruses overcome these nucleotide-based immune mechanisms 🗡️
www.nature.com/articles/s41...

& how viruses overcome these nucleotide-based immune mechanisms 🗡️
www.nature.com/articles/s41...
Excited to share my final work from the @soreklab.bsky.social!
We mined phage dark matter using structural features shared by anti-defense proteins (viral tools that help phages bypass bacterial immunity) to guide discovery.
Found 3 new families targeting immune signaling!
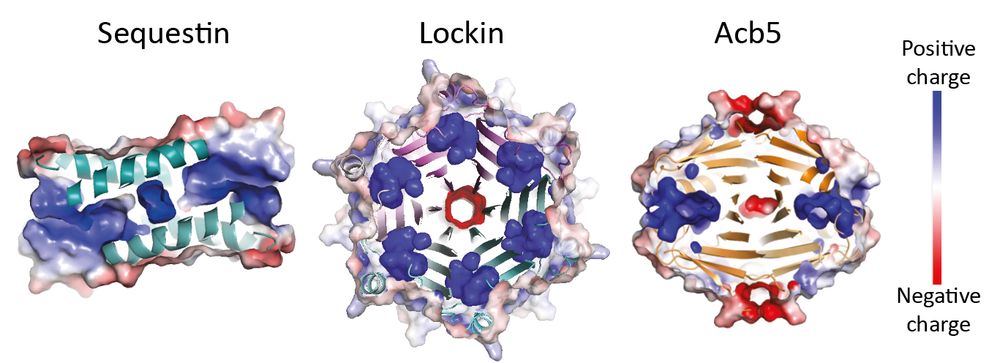
We noticed a pair of genes - a nuclease and a protease - shuffles between antiviral systems. We show how proteolysis activates the nuclease, triggering defense in known and unknown immune contexts.
tinyurl.com/2uwwy4ty

We noticed a pair of genes - a nuclease and a protease - shuffles between antiviral systems. We show how proteolysis activates the nuclease, triggering defense in known and unknown immune contexts.
tinyurl.com/2uwwy4ty


Or do you want to figure out if a phage has a modified genome?
Meet the END-nucleases, an enzyme family that can broadly restrict phages with many diverse modifications. From talented post-doc Wearn-Xin Yee!

Or do you want to figure out if a phage has a modified genome?
Meet the END-nucleases, an enzyme family that can broadly restrict phages with many diverse modifications. From talented post-doc Wearn-Xin Yee!

Check it out:

Check it out:
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
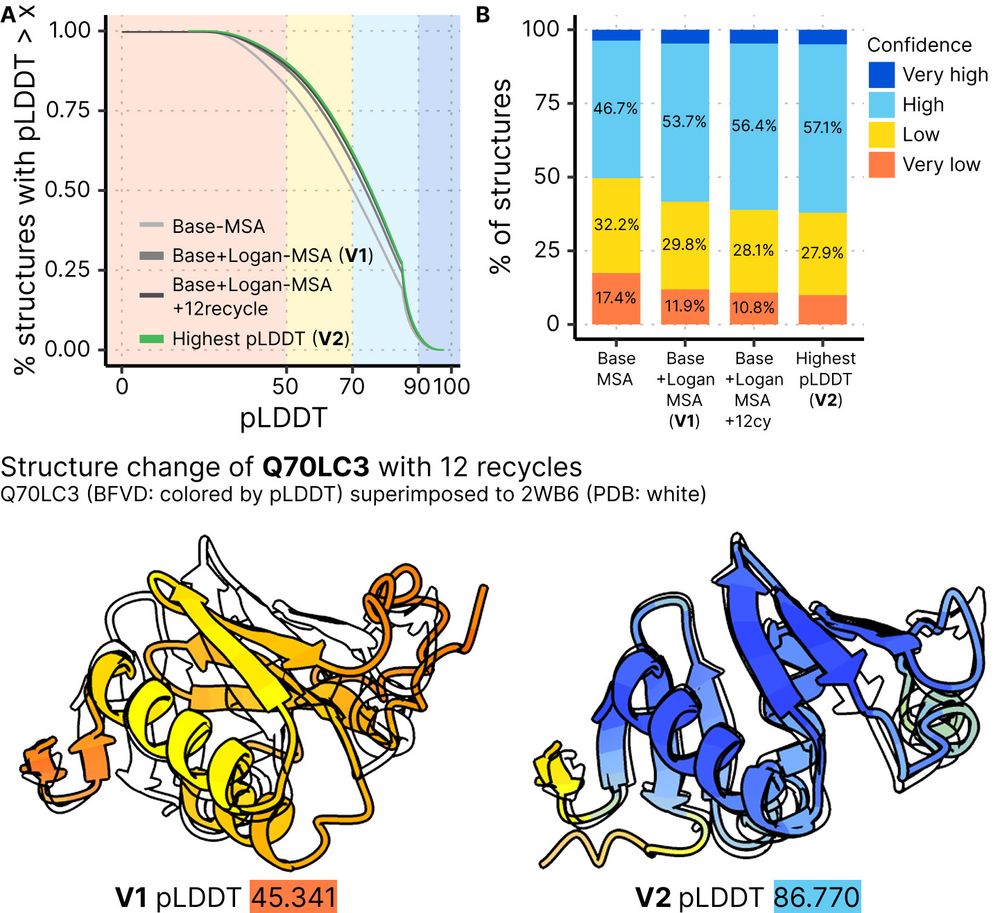
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3


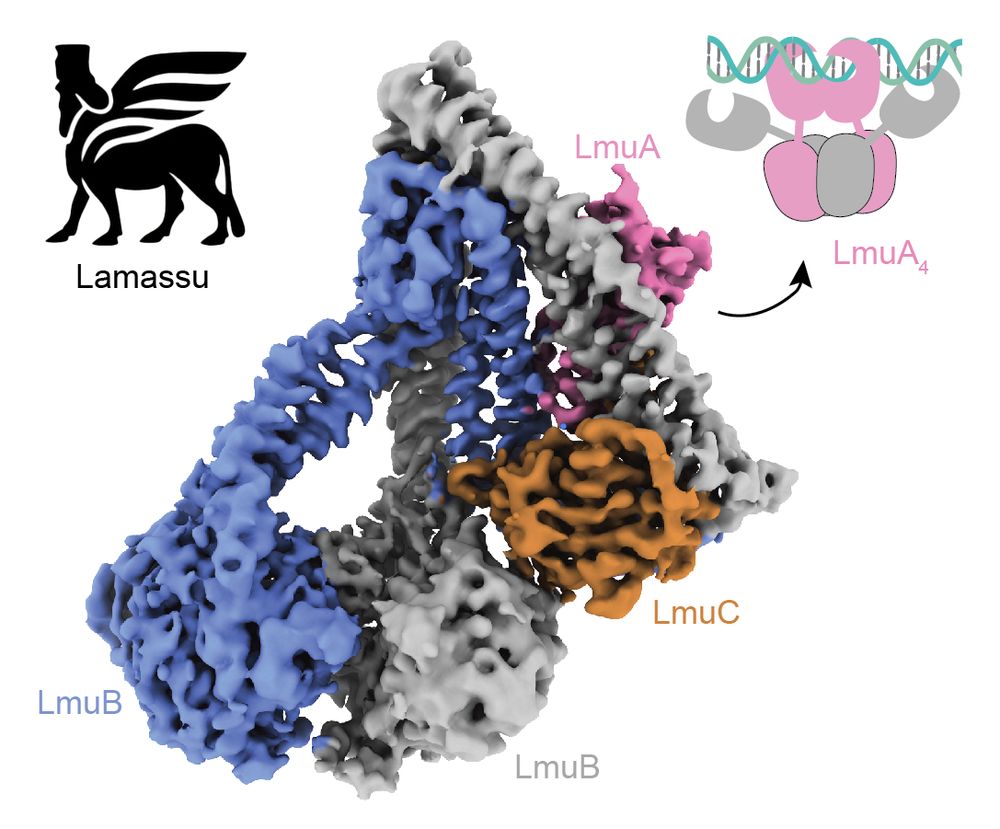
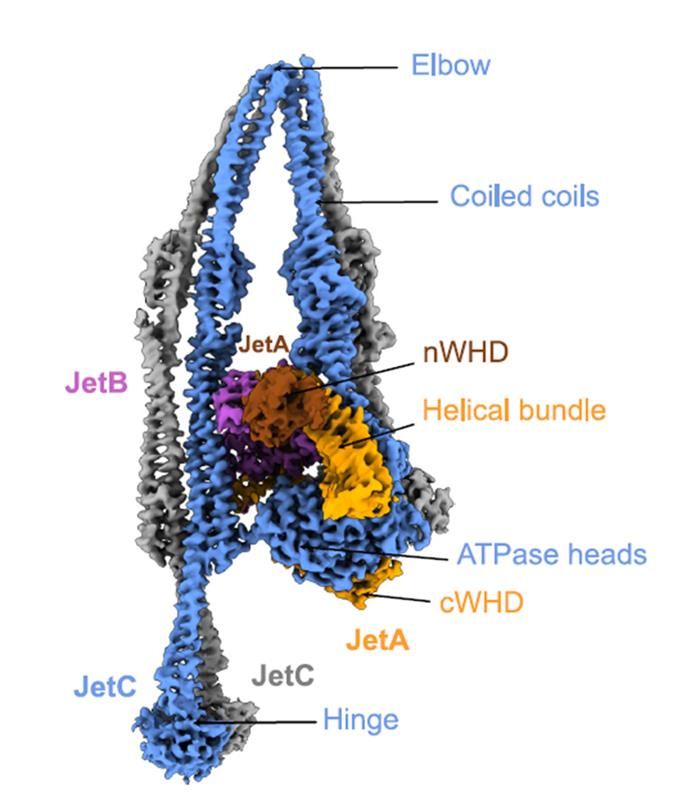
We used functional metagenomics to identify phage defenses in human and soil microbiomes. We scaled these selections while maintaining accuracy, enabling us to examine 9 habitats for defense elements against 7 phages.
www.biorxiv.org/content/10.1...
1/10
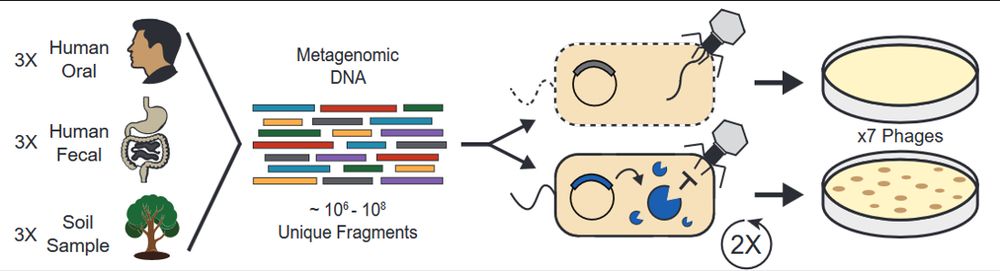
We used functional metagenomics to identify phage defenses in human and soil microbiomes. We scaled these selections while maintaining accuracy, enabling us to examine 9 habitats for defense elements against 7 phages.
www.biorxiv.org/content/10.1...
1/10
We decided to call them TIGR-Tas systems 🐯
We decided to call them TIGR-Tas systems 🐯