🌐https://bfvd.foldseek.com
💾https://bfvd.steineggerlab.workers.dev/
1/3
mirdita.org

mirdita.org
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
It integrates database curation, read QC, taxonomic profiling, and visualization right on your desktop.
No command line, server, or internet required.
Now published in Bioinformatics! 🧵1/5
doi.org/10.1093/bioi...
github.com/steineggerla...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
📄 www.biorxiv.org/content/10.1...
Also check out poster:
B-50 lolalign Sensitive structural alignments by Lasse
B-123 BFVD by Rachel

📄 www.biorxiv.org/content/10.1...
Also check out poster:
B-50 lolalign Sensitive structural alignments by Lasse
B-123 BFVD by Rachel
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social
📄 academic.oup.com/nar/article/...


📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco

For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
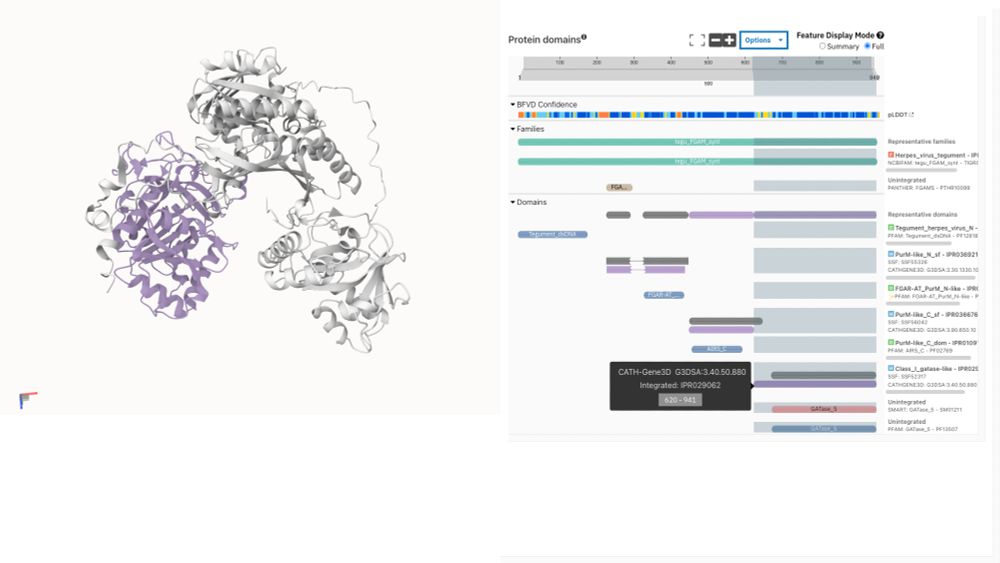
For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
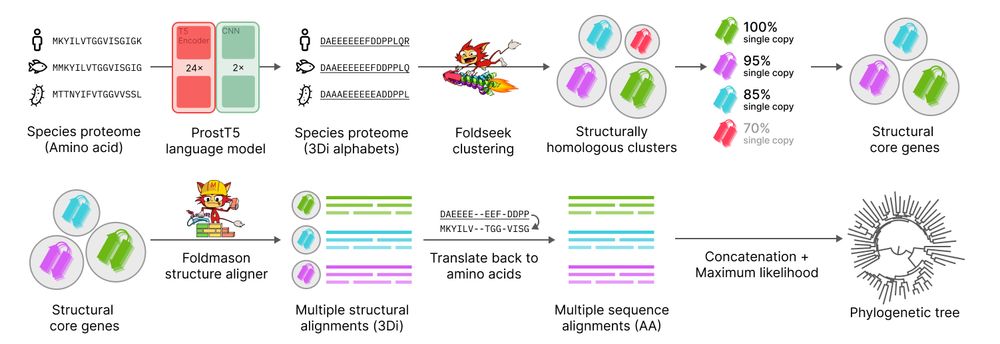
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
🧬🖥️
blog.opentargets.org/an-atlas-of-...

🧬🖥️
blog.opentargets.org/an-atlas-of-...
I'm thrilled to present BFVD at RdRp summit, finally found right place. Thank you for organizers for inviting to this awesome meeting and city.
I would appreciate any feedback and feel free to discuss in-person or cia message. Explore BFVD🧭
Truly FANTASTIC work!
#RdRpSummit2025

I'm thrilled to present BFVD at RdRp summit, finally found right place. Thank you for organizers for inviting to this awesome meeting and city.
I would appreciate any feedback and feel free to discuss in-person or cia message. Explore BFVD🧭
It is Rayan Chikhi!!!
He will introduce us to Logan. You can read the preprint here: tinyurl.com/vx4cykr7
Registrations are still open for #RdRpSummit2025!

It is Rayan Chikhi!!!
He will introduce us to Logan. You can read the preprint here: tinyurl.com/vx4cykr7
Registrations are still open for #RdRpSummit2025!
The registrations for on-site & remote participation are still open! More info: RdRp.io
#RdRpSummit2025

The registrations for on-site & remote participation are still open! More info: RdRp.io
#RdRpSummit2025
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
📄 academic.oup.com/nar/article/...
🌐 bfvd.foldseek.com
#RECOMB2025

📄 academic.oup.com/nar/article/...
🌐 bfvd.foldseek.com
#RECOMB2025
✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕




✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕

