
📃 doi.org/10.64898/2026.01.26.701724
📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...

📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...
📃 doi.org/10.64898/2026.01.26.701724
📃 doi.org/10.64898/2026.01.26.701724
mirdita.org

mirdita.org
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
It’s faster, more accurate, and ready for thousands of genomes
Let’s break it down (1/10)
github.com/OrthoFinder/...
www.biorxiv.org/content/10.1...
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco

Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
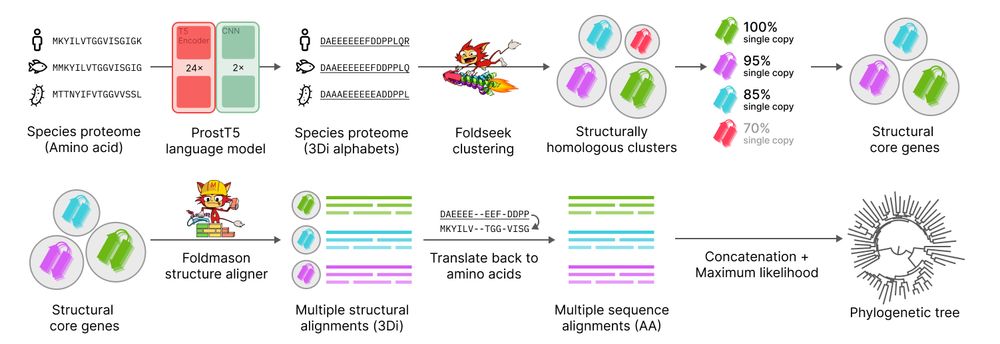
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
(Most credit to Minh Bui and @roblanfear.bsky.social and their labs)
ecoevorxiv.org/repository/v...
(Most credit to Minh Bui and @roblanfear.bsky.social and their labs)
ecoevorxiv.org/repository/v...
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
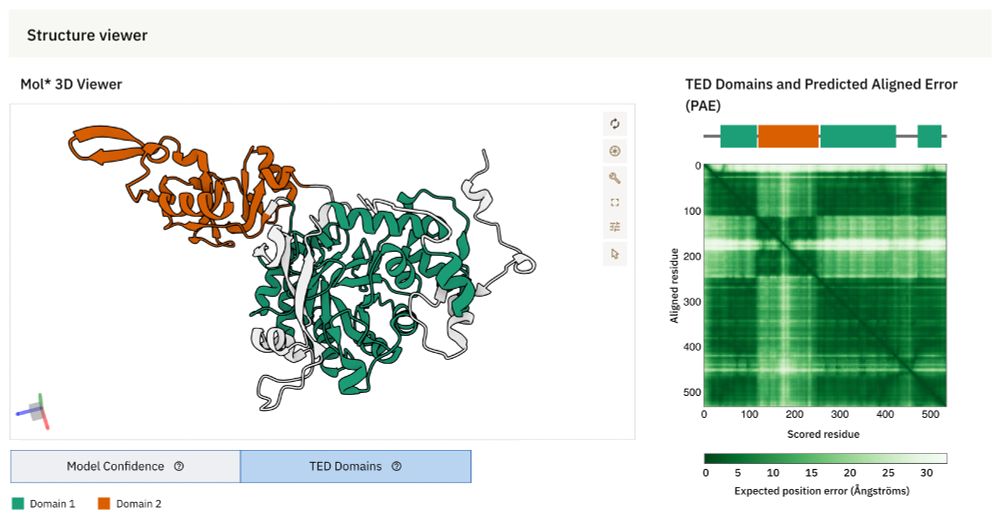
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
www.sib.swiss/news/sib-hel...
www.sib.swiss/news/sib-hel...
📄 doi.org/10.1093/nsr/...
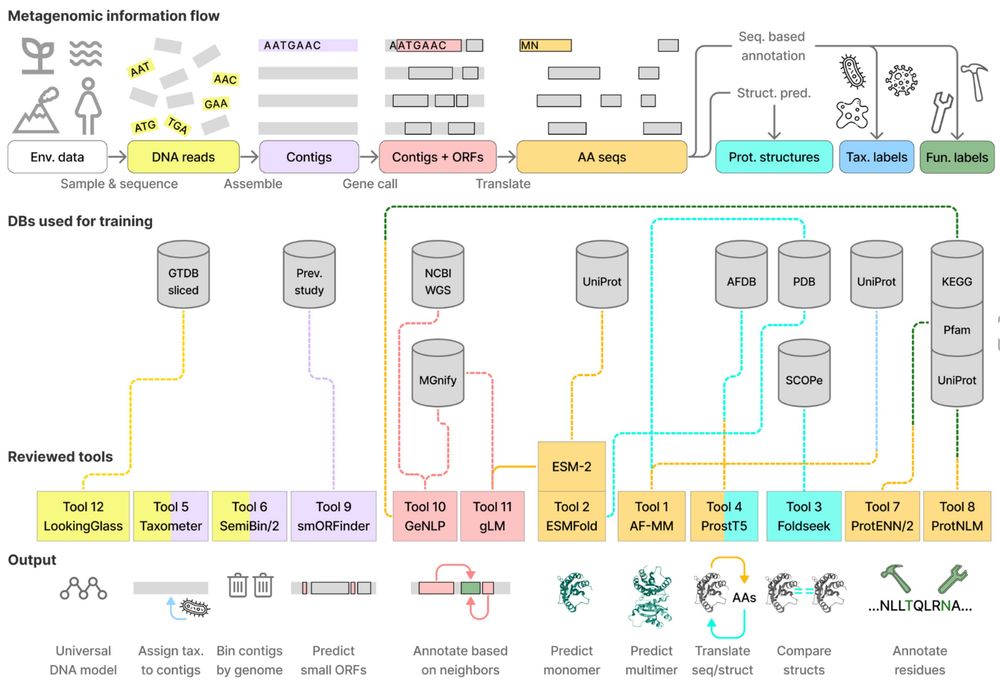
📄 doi.org/10.1093/nsr/...
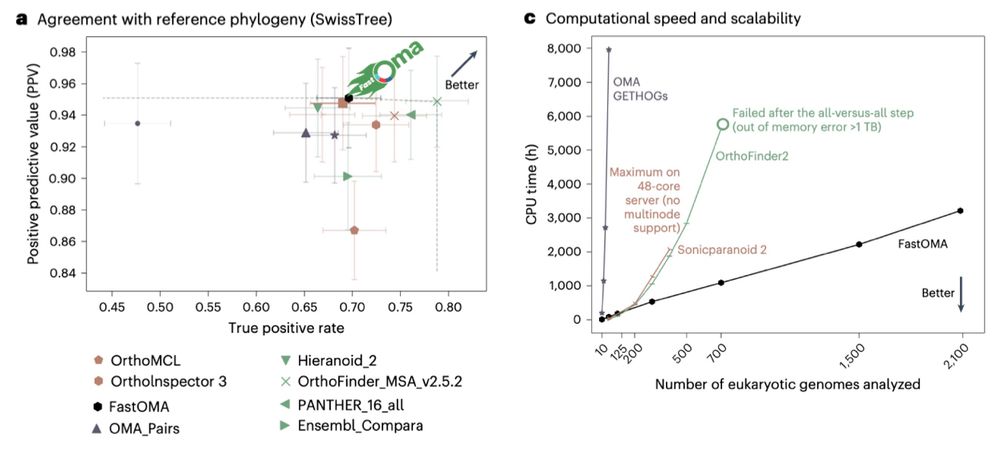
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
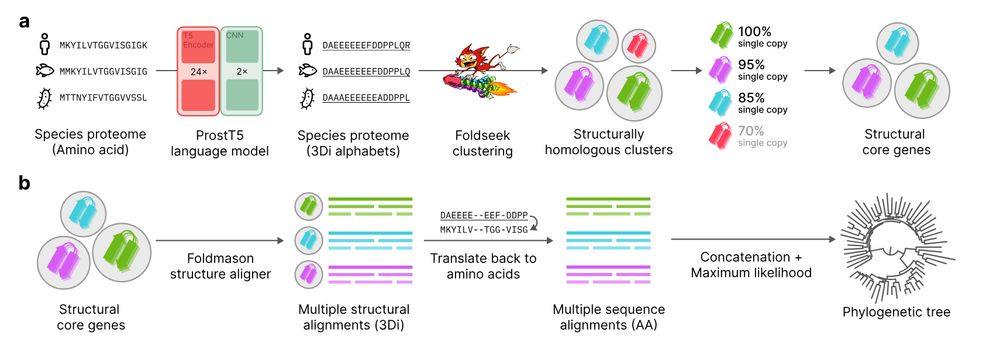
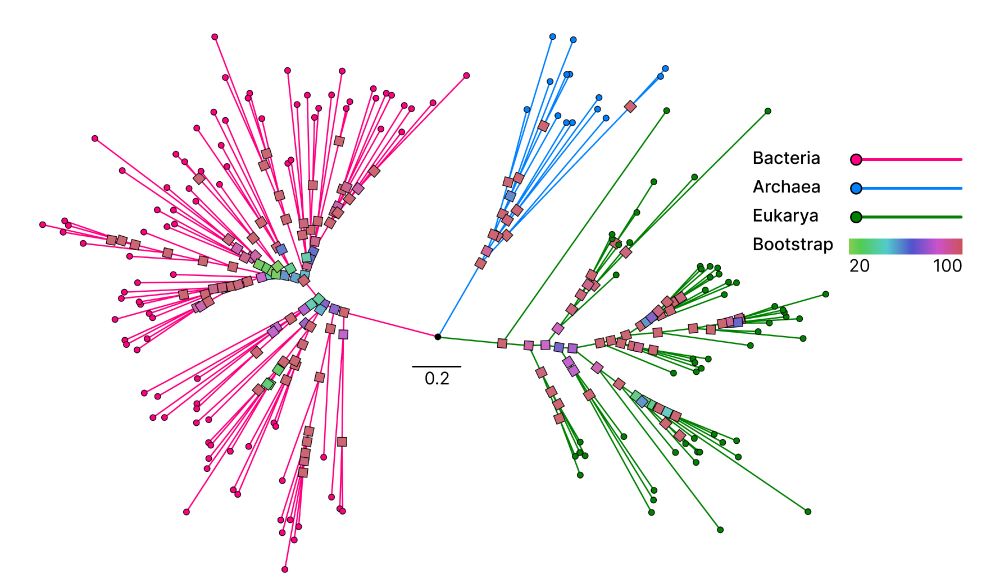
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
A Place of Joy.
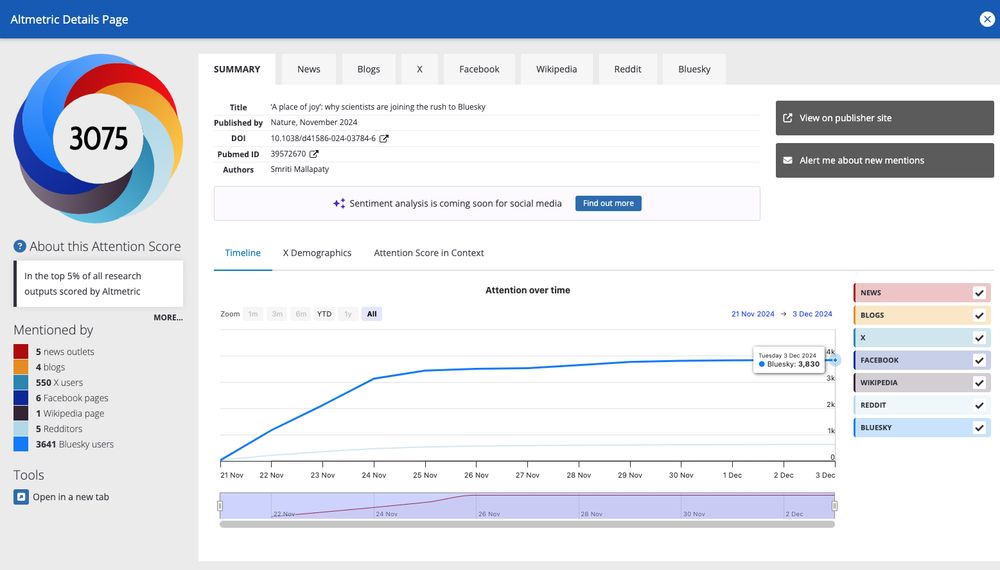
🔗 go.bsky.app/VJhXcSs
🔗 go.bsky.app/VJhXcSs
📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶

📄 biorxiv.org/content/10.1...
💾 mmseqs.com and 🐍Bioconda 🖥️🧬🧶
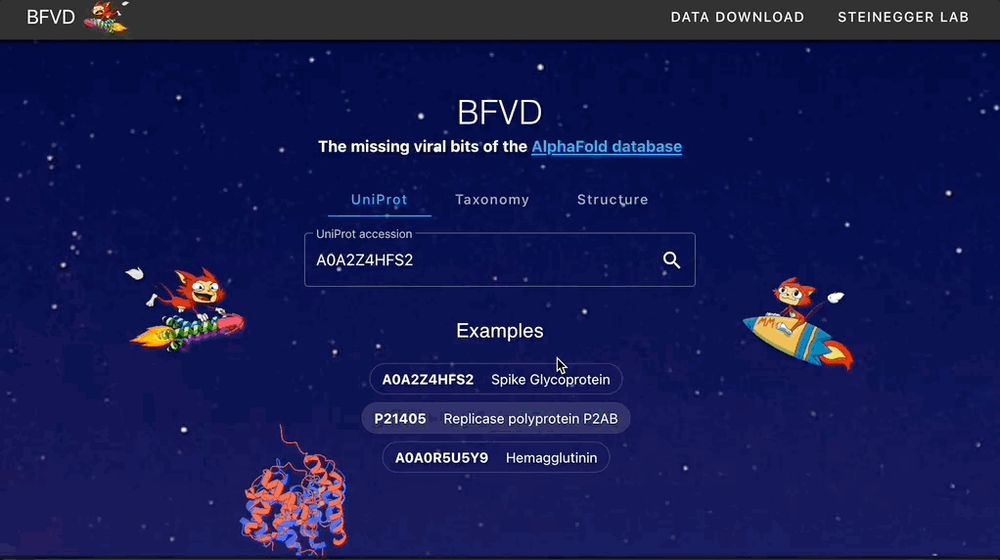
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...
🌐 bfvd.foldseek.com
💾 bfvd.steineggerlab.workers.dev
📄 academic.oup.com/nar/advance-...

