
Julia programming.
https://diegozea.github.io/
#JuliaLang #SciML #Python #MATLAB #ScientificComputing #DataScience #Cheatsheet #Engineering

#JuliaLang #SciML #Python #MATLAB #ScientificComputing #DataScience #Cheatsheet #Engineering

📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
The application deadline for the PhD student position has been extended to July 4th. If you haven’t applied yet, there’s still time! 👇
as part of the ANR SPPICES project! 🚀
📅 Start: 1 Sept 2025
⏳ Deadline: 23 June 2025
👉Apply now: emploi.cnrs.fr/Offres/Docto...
The application deadline for the PhD student position has been extended to July 4th. If you haven’t applied yet, there’s still time! 👇
🛠️ Develop #JuliaLang & Python tools to model structural and evolutionary features of IDPs
📅 Start: 1 Sept 2025
🎓 3+ years of higher education (Master’s or engineering diploma preferred)
👉 Apply now: emploi.cnrs.fr/Offres/CDD/U...
🛠️ Develop #JuliaLang & Python tools to model structural and evolutionary features of IDPs
📅 Start: 1 Sept 2025
🎓 3+ years of higher education (Master’s or engineering diploma preferred)
👉 Apply now: emploi.cnrs.fr/Offres/CDD/U...
as part of the ANR SPPICES project! 🚀
📅 Start: 1 Sept 2025
⏳ Deadline: 23 June 2025
👉Apply now: emploi.cnrs.fr/Offres/Docto...
as part of the ANR SPPICES project! 🚀
📅 Start: 1 Sept 2025
⏳ Deadline: 23 June 2025
👉Apply now: emploi.cnrs.fr/Offres/Docto...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
@msftresearch.bsky.social
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...

@msftresearch.bsky.social

doi.org/10.1371/jour...
🧬🧶
doi.org/10.1371/jour...
🧬🧶

go.bsky.app/Hv4RKzM
What other starter packs would you want to see? I'm in health research, mathematics, and open source/science. 🧪🛟🩺 📊
go.bsky.app/Hv4RKzM
What other starter packs would you want to see? I'm in health research, mathematics, and open source/science. 🧪🛟🩺 📊
AI: go.bsky.app/SipA7it
RL: go.bsky.app/3WPHcHg
Women in AI: go.bsky.app/LaGDpqg
NLP: go.bsky.app/SngwGeS
AI and news: go.bsky.app/5sFqVNS
You can also search all starter packs here: blueskydirectory.com/starter-pack...
AI: go.bsky.app/SipA7it
RL: go.bsky.app/3WPHcHg
Women in AI: go.bsky.app/LaGDpqg
NLP: go.bsky.app/SngwGeS
AI and news: go.bsky.app/5sFqVNS
You can also search all starter packs here: blueskydirectory.com/starter-pack...
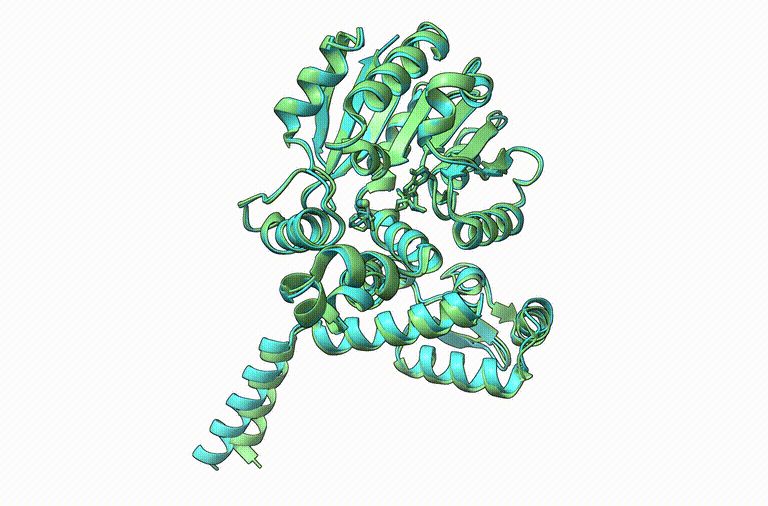
🔄Share to help people finding its topic/field
- STRUCTURAL BIOLOGY
Structural Biology 1: bsky.app/starter-pack...
Structural Biology 2: bsky.app/starter-pack...
Crystallography: bsky.app/starter-pack...
Synchrotron&CryoEM: bsky.app/starter-pack...

🔄Share to help people finding its topic/field
- STRUCTURAL BIOLOGY
Structural Biology 1: bsky.app/starter-pack...
Structural Biology 2: bsky.app/starter-pack...
Crystallography: bsky.app/starter-pack...
Synchrotron&CryoEM: bsky.app/starter-pack...
1/8

1/8
colab.research.google.com/drive/17e0pD...

colab.research.google.com/drive/17e0pD...
rdcu.be/dwlLa
Work done with Hélène Bret and Raphael Guerois and for the revised version with great contributions from @diegozea.bsky.social and Jinmei Gao.
🧬🧶
rdcu.be/dwlLa
Work done with Hélène Bret and Raphael Guerois and for the revised version with great contributions from @diegozea.bsky.social and Jinmei Gao.
🧬🧶
Everyone is welcome!

Everyone is welcome!

www.youtube.com/watch?v=JD8H...

www.youtube.com/watch?v=JD8H...

