EMBO Fellow doing Computational Proteomics
Nesvi lab -> Özlü lab
📍İstanbul
https://danny.bio

What was previously reported to have little global impact now is reported to explain up to half of the variation in how protein levels are controlled. Protein degradation matters!
shorturl.at/5wPiS

What was previously reported to have little global impact now is reported to explain up to half of the variation in how protein levels are controlled. Protein degradation matters!
shorturl.at/5wPiS
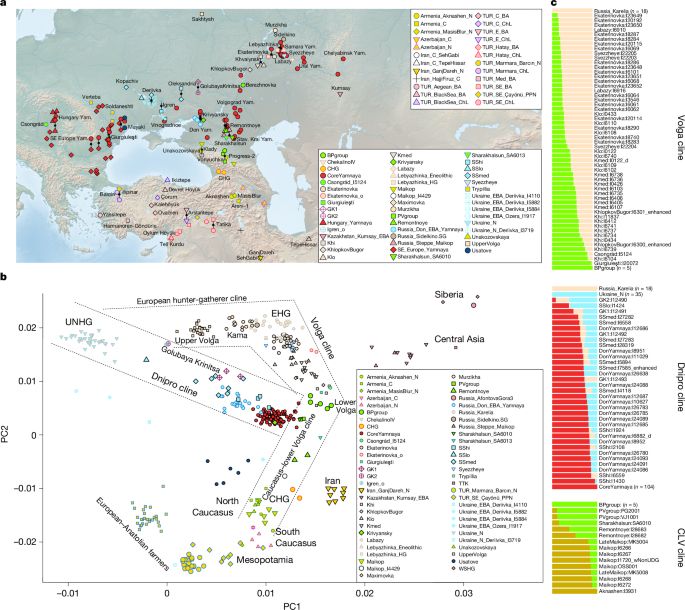
www.nature.com/articles/s41...
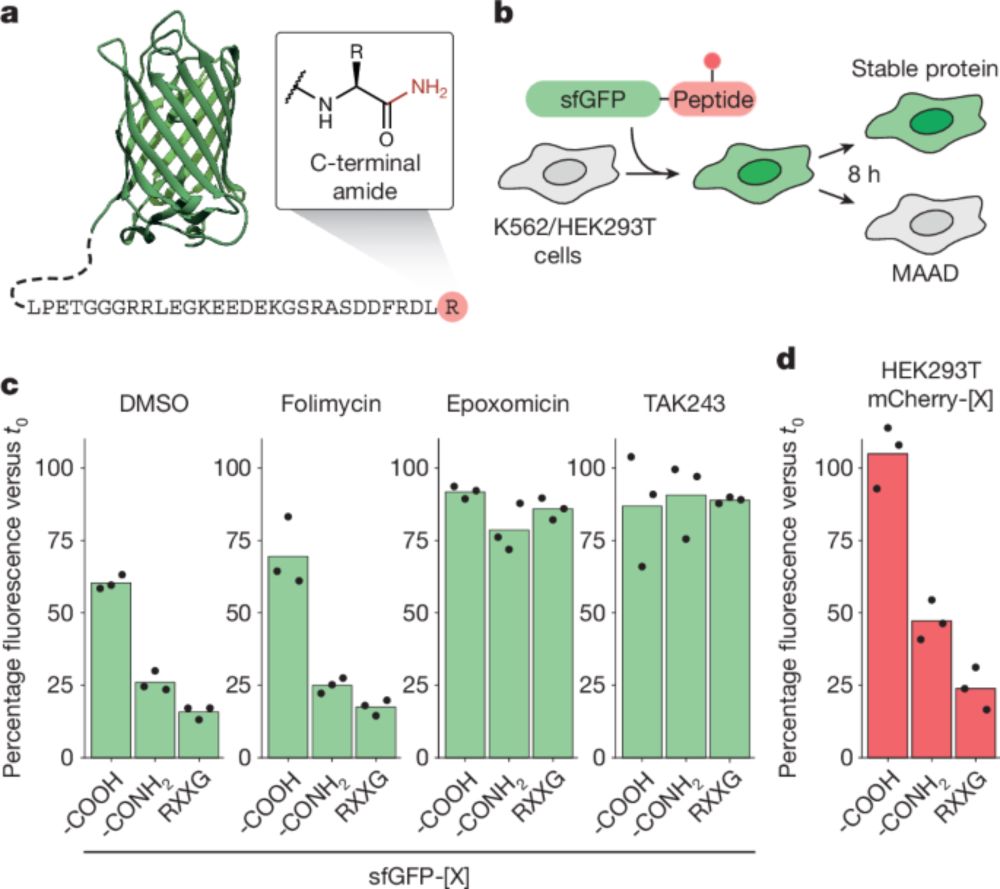
www.nature.com/articles/s41...
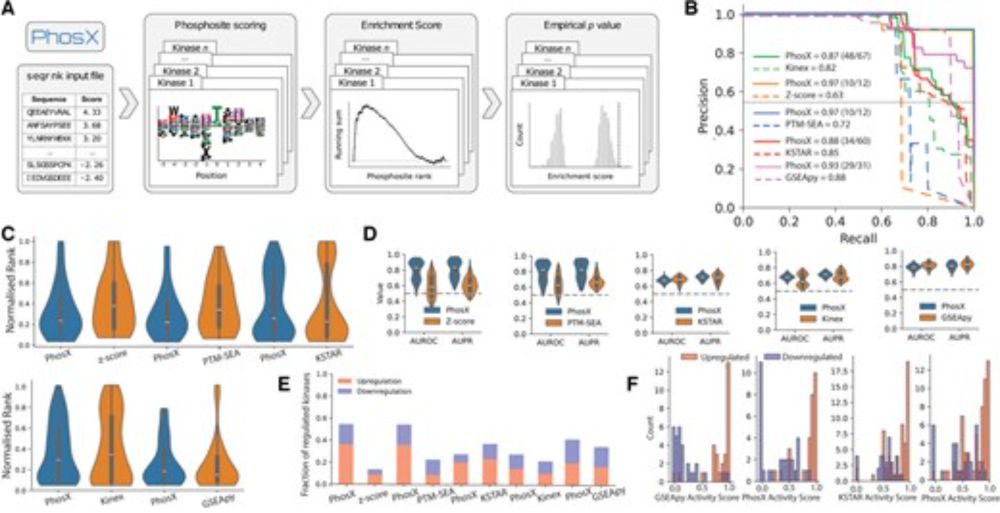
Enter PhosX, a new tool transforming the way researchers analyse and interpret kinase activity.
Read the full article: academic.oup.com/bioinformati...
@alussana.bsky.social @epetsalaki.bsky.social

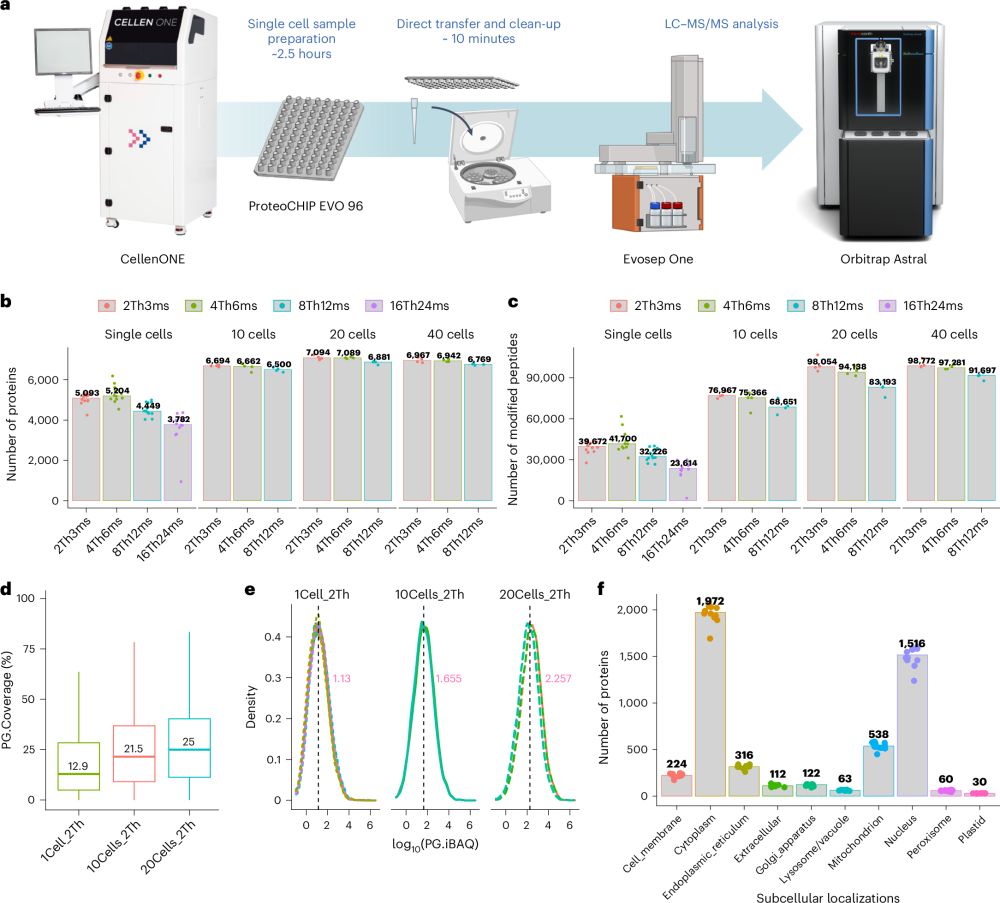
📄 doi.org/10.1038/s414... (1/6)
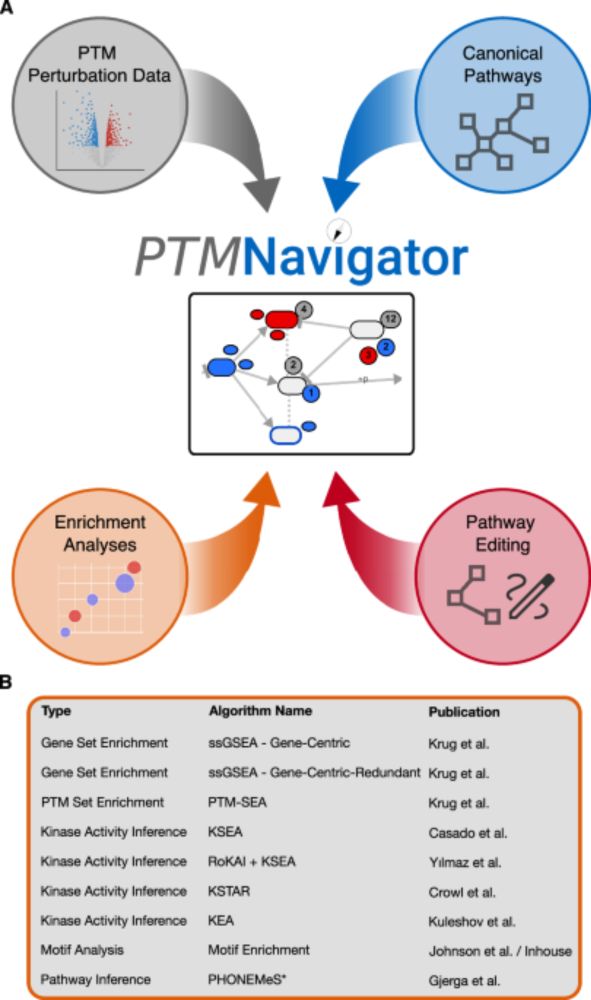
📄 doi.org/10.1038/s414... (1/6)
www.linkedin.com/posts/chrisd...

www.linkedin.com/posts/chrisd...
mathstodon.xyz/@11011110/11...
www.nature.com/articles/s41...

www.nature.com/articles/s41...



www.nature.com/articles/s41...
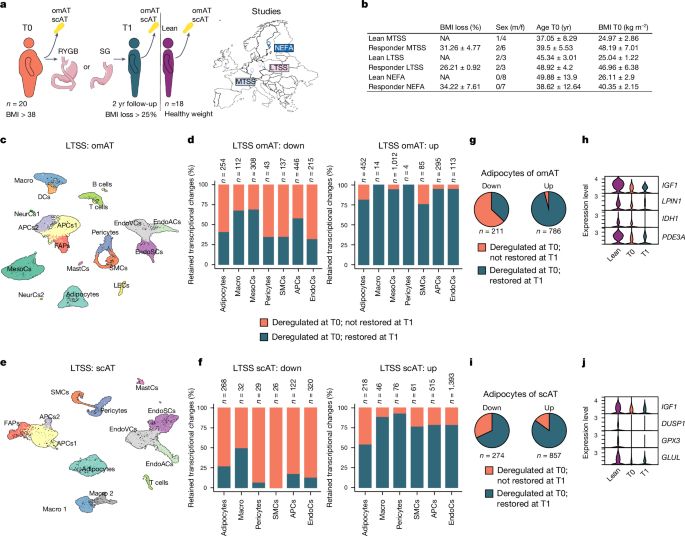
www.nature.com/articles/s41...
nvidia.github.io/bionemo-fram...
nvidia.github.io/bionemo-fram...