Working with the Functional Metabolomics Lab on developing mass spec tools to understand microbial communities.
www.functional-metabolomics.com
#secmet #lassopeptides #syntheticbiology

#secmet #lassopeptides #syntheticbiology





#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...

#phylogeny #secmet #bioinformatics
www.biorxiv.org/content/10.1...
If you want to check it out, you can find our preprint here:
doi.org/10.26434/che...

If you want to check it out, you can find our preprint here:
doi.org/10.26434/che...
www.nature.com/articles/s41...
Behind the paper story:
go.nature.com/45ljV4d
www.nature.com/articles/s41...
Behind the paper story:
go.nature.com/45ljV4d
Thanks so much @simonsfoundation.org
We can’t wait to get started!

Thanks so much @simonsfoundation.org
We can’t wait to get started!
Big congratulations to Shane Farrell and everybody involved! ✌️✌️✌️
Learn more in this week's issue of Science: scim.ag/4dwzl8j

Big congratulations to Shane Farrell and everybody involved! ✌️✌️✌️
Our CMFI Cluster of Excellence @unituebingen.bsky.social receives funding extension for the next seven years.
Spokesperson Andreas Peschel @andreaspeschel.bsky.social: "We can now advance our research into resistance mechanisms and new antimicrobial agents!"
shorturl.at/rcK1A
Our CMFI Cluster of Excellence @unituebingen.bsky.social receives funding extension for the next seven years.
Spokesperson Andreas Peschel @andreaspeschel.bsky.social: "We can now advance our research into resistance mechanisms and new antimicrobial agents!"
shorturl.at/rcK1A
@iocbprague.bsky.social! Both new and advanced users are learning about the latest mzmine features from our amazing instructors @ansgarkorf.bsky.social, Josh Smith,
@titodamiani.bsky.social, and @roman-bushuiev.bsky.social.

@iocbprague.bsky.social! Both new and advanced users are learning about the latest mzmine features from our amazing instructors @ansgarkorf.bsky.social, Josh Smith,
@titodamiani.bsky.social, and @roman-bushuiev.bsky.social.
www.biorxiv.org/content/10.1...
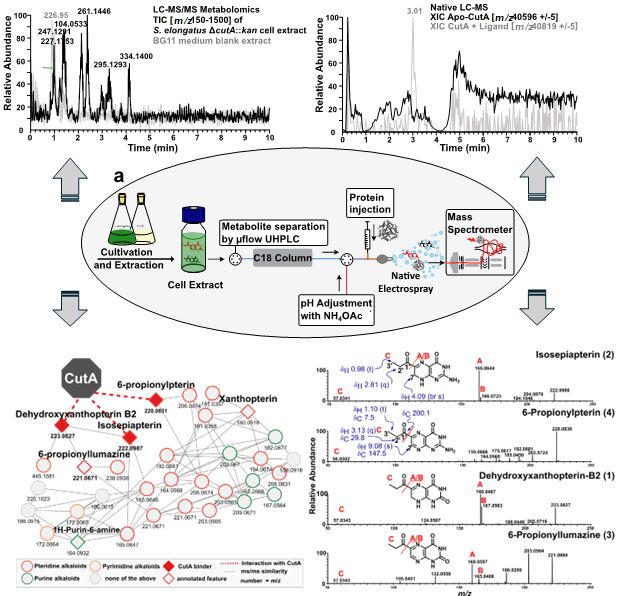
www.biorxiv.org/content/10.1...
doi.org/10.1021/acs....
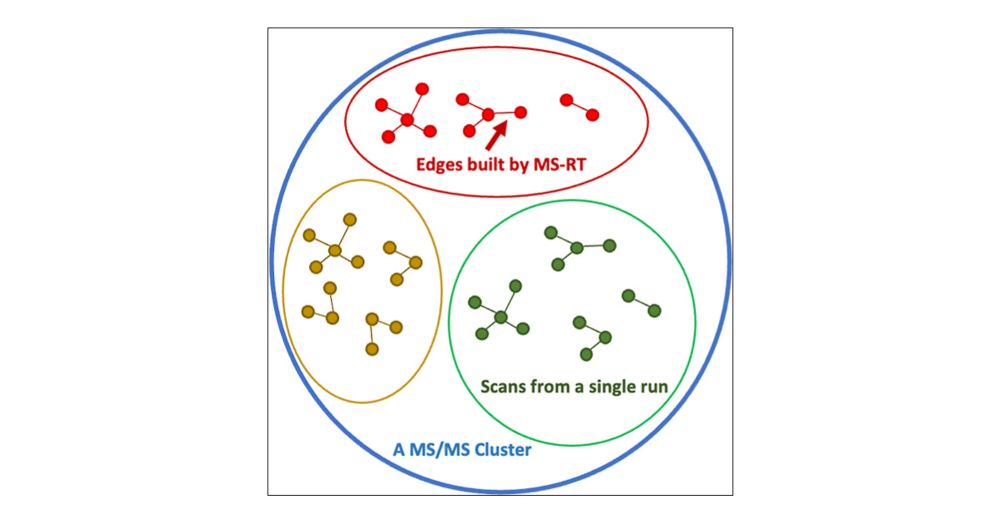
doi.org/10.1021/acs....
By reanalyzing data from 30,000 authentic standards @yelabiead.bsky.social @adafede.bsky.social @pieterdorrestein.bsky.social et al. show that ISFs are substantially less that what Giera et al. reported 👌

By reanalyzing data from 30,000 authentic standards @yelabiead.bsky.social @adafede.bsky.social @pieterdorrestein.bsky.social et al. show that ISFs are substantially less that what Giera et al. reported 👌
Sign up:
tinyurl.com/NTMS2025

doi.org/10.1007/s113...
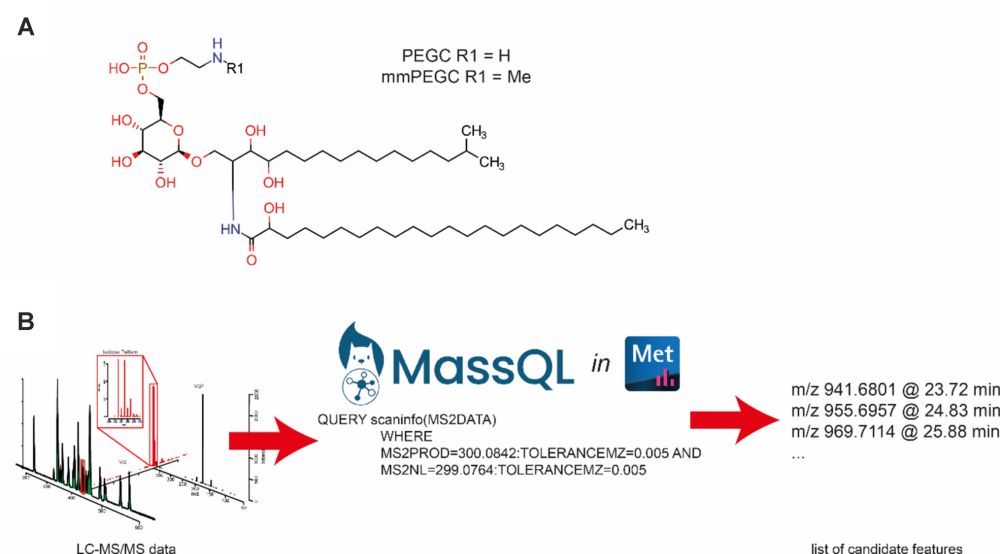
doi.org/10.1007/s113...
Don’t forget to join us for our first webinar of the year:
“Learning From Repository-Scale Untargeted Metabolomics Data”
📅 Date: 22 January
🕒 Time: 3 PM UTC
#MetabolomicsSociety #MetSoc #Metabolomics #ECR #TeamMassSpec #EMNMetSoc
On 22 January at 3pm UTC, we will host Dr Wout Bittremieux who will deliver a talk on an approach to untargeted metabolomics data annotation!
✒️ Register here for free: tinyurl.com/2v654vp9
We look forward to seeing you there 👋

Don’t forget to join us for our first webinar of the year:
“Learning From Repository-Scale Untargeted Metabolomics Data”
📅 Date: 22 January
🕒 Time: 3 PM UTC
#MetabolomicsSociety #MetSoc #Metabolomics #ECR #TeamMassSpec #EMNMetSoc




The coolest feature is a new AI-based charge assignment for isotopic resolution data. It's performing a little better than Thrash/Xtract but way faster. Try out IsoDec if you are doing top-down!
github.com/michaelmarty...
The coolest feature is a new AI-based charge assignment for isotopic resolution data. It's performing a little better than Thrash/Xtract but way faster. Try out IsoDec if you are doing top-down!
github.com/michaelmarty...

A limited number of supporting grants will be given to early career researchers and #ICTC13 participants from lower income countries.
Don't miss the opportunity to submit an abstract and apply for an #ICTC13 grant !
Deadline: 20 December
More: ictc13.gr/grants/
A limited number of supporting grants will be given to early career researchers and #ICTC13 participants from lower income countries.
Don't miss the opportunity to submit an abstract and apply for an #ICTC13 grant !
Deadline: 20 December
More: ictc13.gr/grants/