thomas-a-neil.github.io
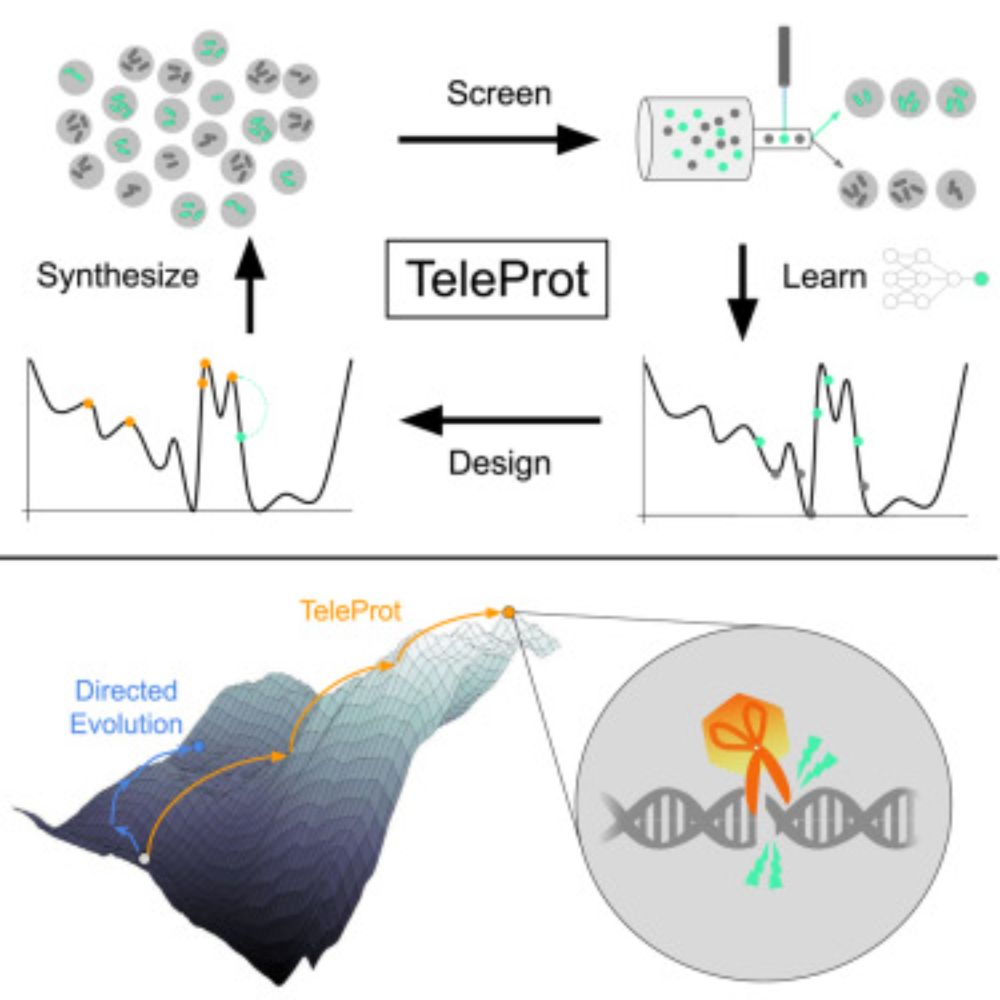
Fun collaboration between Google X, DeepMind, and Triplebar that we hope can be a template for integrating ML and high throughput screening in protein engineering
Day 1/31
Prompt MUSTACHE
Pdb 2QZI
Let’s start with something fun:
Mr. Potato head’s ‘stache is made of Androgen Receptor that binds testosterone and helps maintain his male phenotype
Next prompt: WEAVE
suggestions?

Day 1/31
Prompt MUSTACHE
Pdb 2QZI
Let’s start with something fun:
Mr. Potato head’s ‘stache is made of Androgen Receptor that binds testosterone and helps maintain his male phenotype
Next prompt: WEAVE
suggestions?
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)

t.co/qCZERPUMPF
t.co/qCZERPUMPF
🌍 Design a PETase - real-world impact on bioremediation!
🧬 Sponsored DNA synthesis and functional screening - no need for a lab!
🤖 Sponsored ESM inference through @evolutionaryscale.bsky.social Forge - if GPUs are a barrier!
🏆 Winners get published and win up to $15K!
PETase can degrade PET, but isn’t ready for industrial-scale waste. The challenge: design an improved variant that can change that.
Register by Oct 17 alignbio.org/protein-engi...

🌍 Design a PETase - real-world impact on bioremediation!
🧬 Sponsored DNA synthesis and functional screening - no need for a lab!
🤖 Sponsored ESM inference through @evolutionaryscale.bsky.social Forge - if GPUs are a barrier!
🏆 Winners get published and win up to $15K!
PETase can degrade PET, but isn’t ready for industrial-scale waste. The challenge: design an improved variant that can change that.
Register by Oct 17 alignbio.org/protein-engi...

PETase can degrade PET, but isn’t ready for industrial-scale waste. The challenge: design an improved variant that can change that.
Register by Oct 17 alignbio.org/protein-engi...
- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
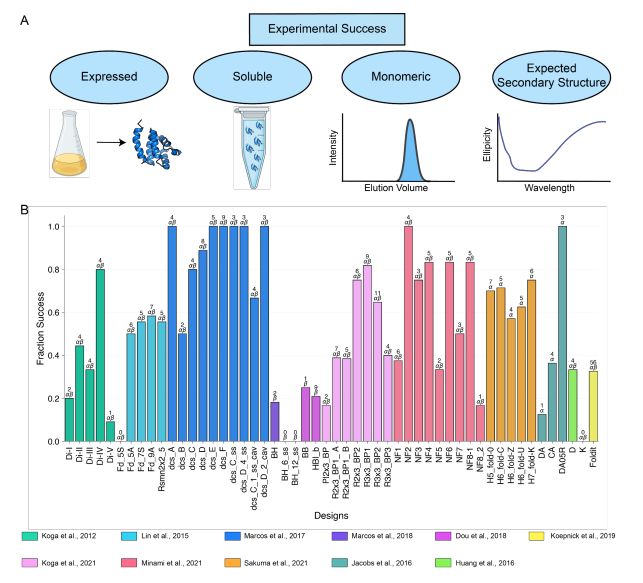



- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
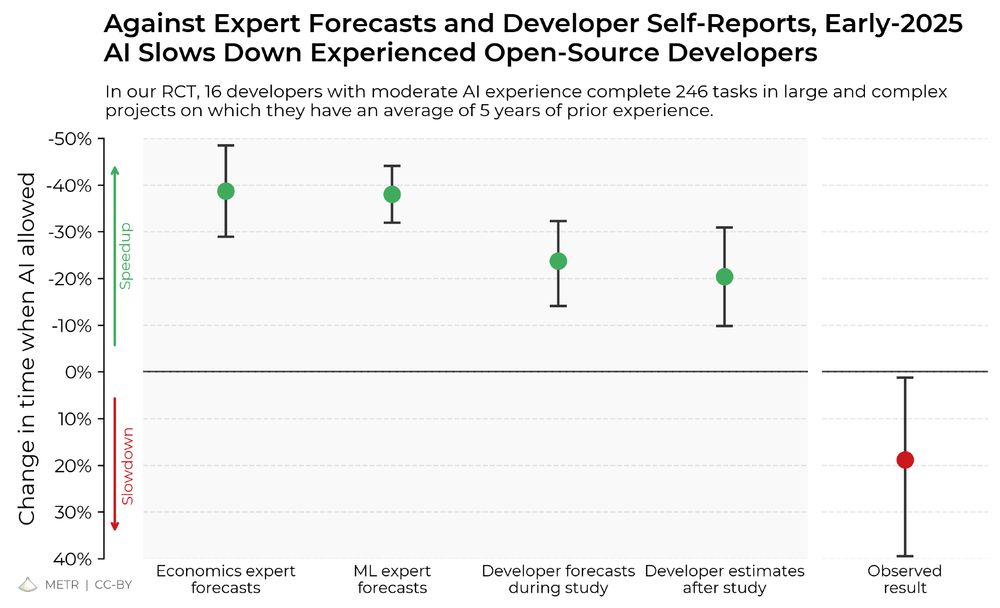
The results surprised us: Developers thought they were 20% faster with AI tools, but they were actually 19% slower when they had access to AI than when they didn't.
🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience

🚀 Announcing the 2025 Protein Engineering Tournament.
This year’s challenge: design PETase enzymes, which degrade the type of plastic in bottles. Can AI-guided protein design help solve the climate crisis? Let’s find out! ⬇️
#AIforBiology #ClimateTech #ProteinEngineering #OpenScience
Get started using ESMC to predict protein function and ESM3 to generate new enzymes here: github.com/evolutionary...
Get started using ESMC to predict protein function and ESM3 to generate new enzymes here: github.com/evolutionary...
Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!

Paper: biorxiv.org/content/10.1...
Sign up on our website for zoom links!

We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
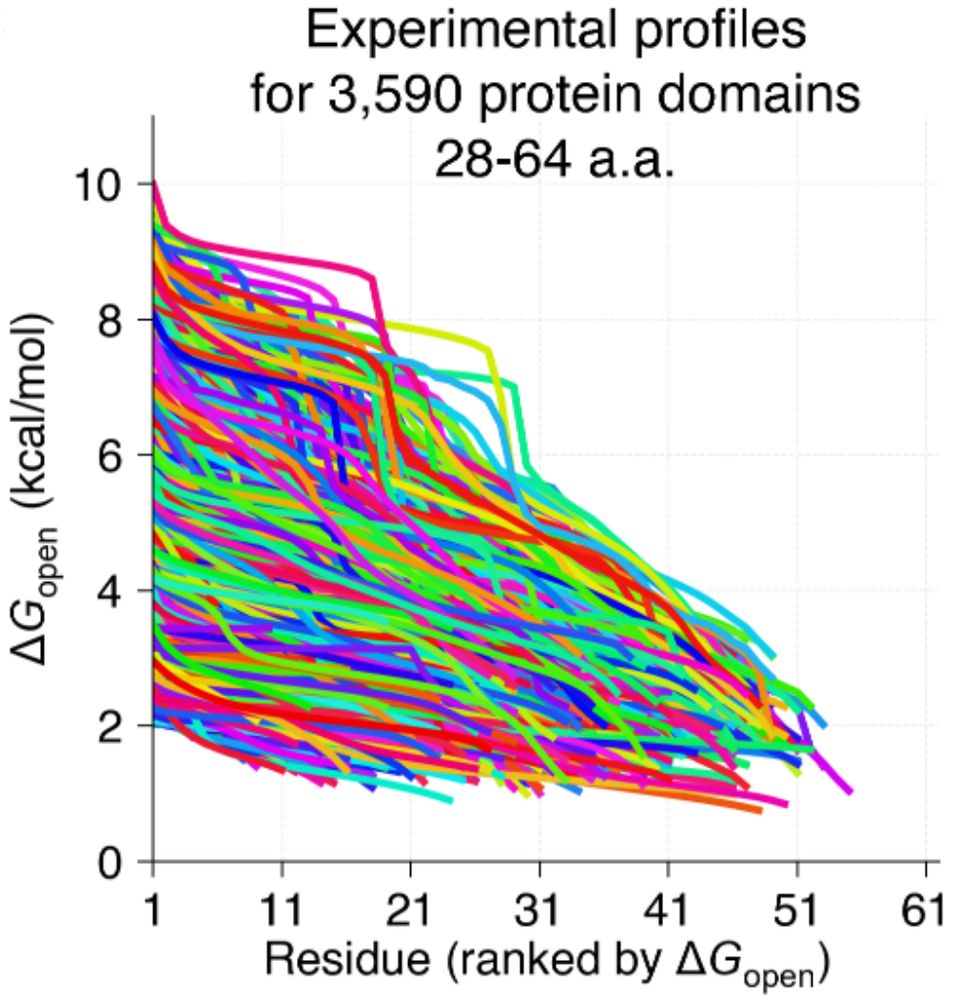
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
Happy to have played a small part in this work, where Chase developed a method for precision library construction at scale, with per-gene costs as low as $1.50.
@philromero.bsky.social

Happy to have played a small part in this work, where Chase developed a method for precision library construction at scale, with per-gene costs as low as $1.50.
@philromero.bsky.social
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Today w @ginaelnesr.bsky.social, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained w them: Dyna-1.
📝: rb.gy/de5axp
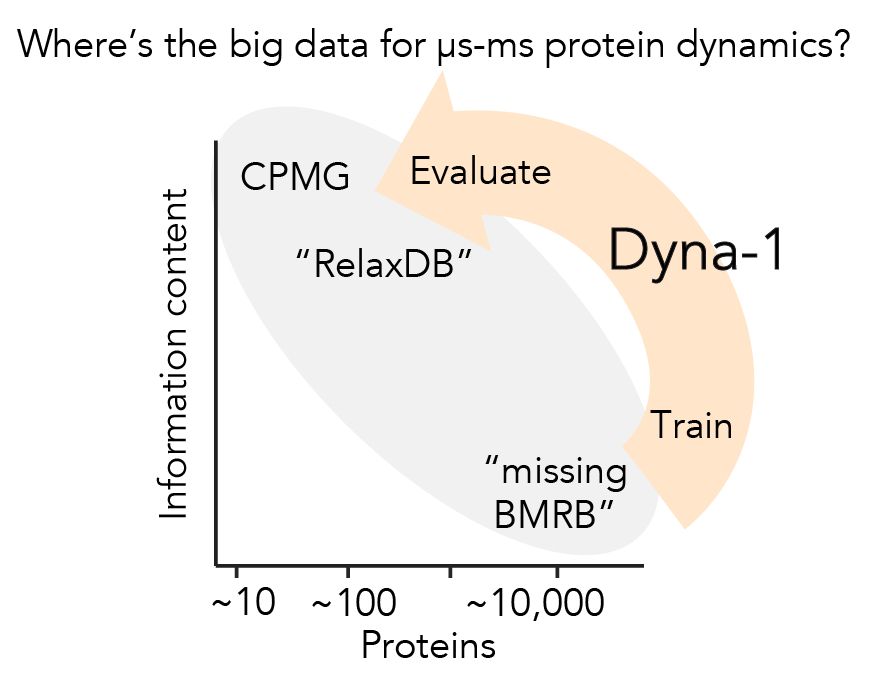
Today w @ginaelnesr.bsky.social, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained w them: Dyna-1.
📝: rb.gy/de5axp
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
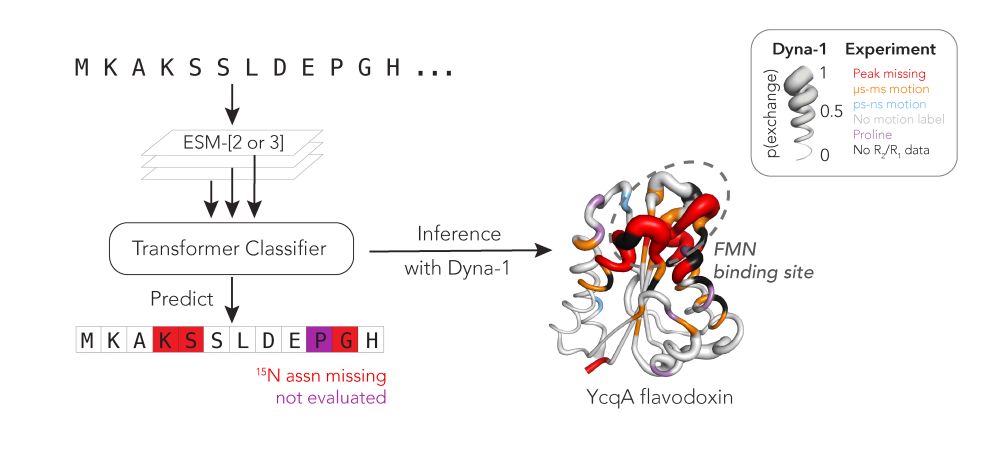
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
Fun collaboration between Google X, DeepMind, and Triplebar that we hope can be a template for integrating ML and high throughput screening in protein engineering

Fun collaboration between Google X, DeepMind, and Triplebar that we hope can be a template for integrating ML and high throughput screening in protein engineering
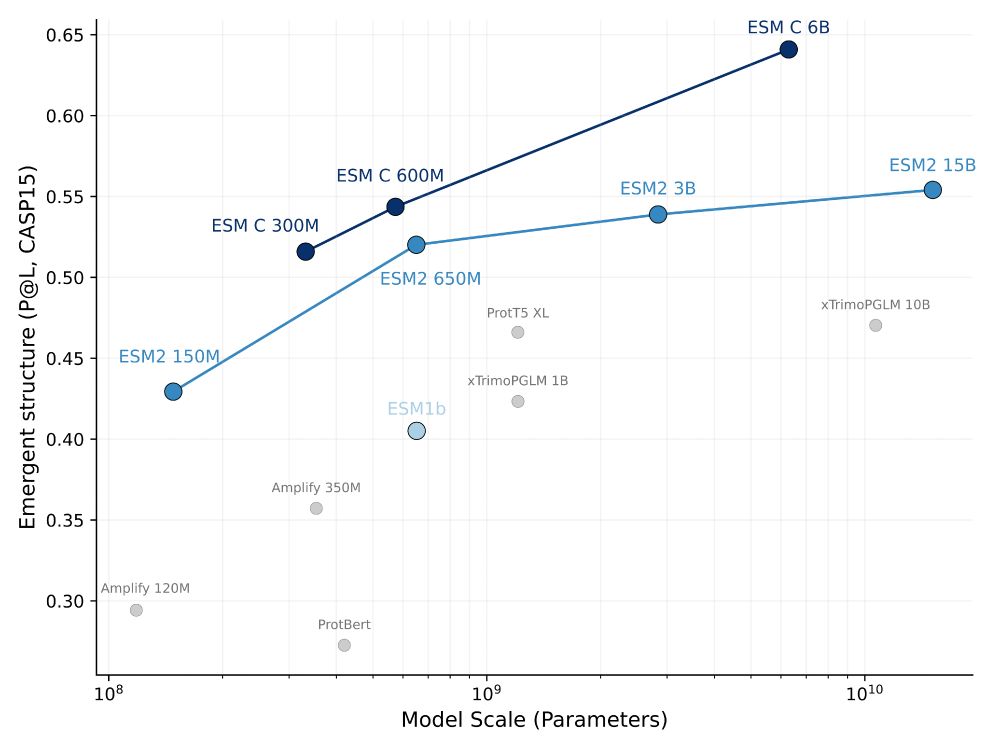
1/7
openreview.net/forum?id=H7m...

1/7
openreview.net/forum?id=H7m...

