
doi.org/10.1038/s415...

doi.org/10.1038/s415...
sites.google.com/view/codonus...



Great work by Linglan Fang and Wenrui Zhong in our lab!
doi.org/10.1021/jacs.5c11900


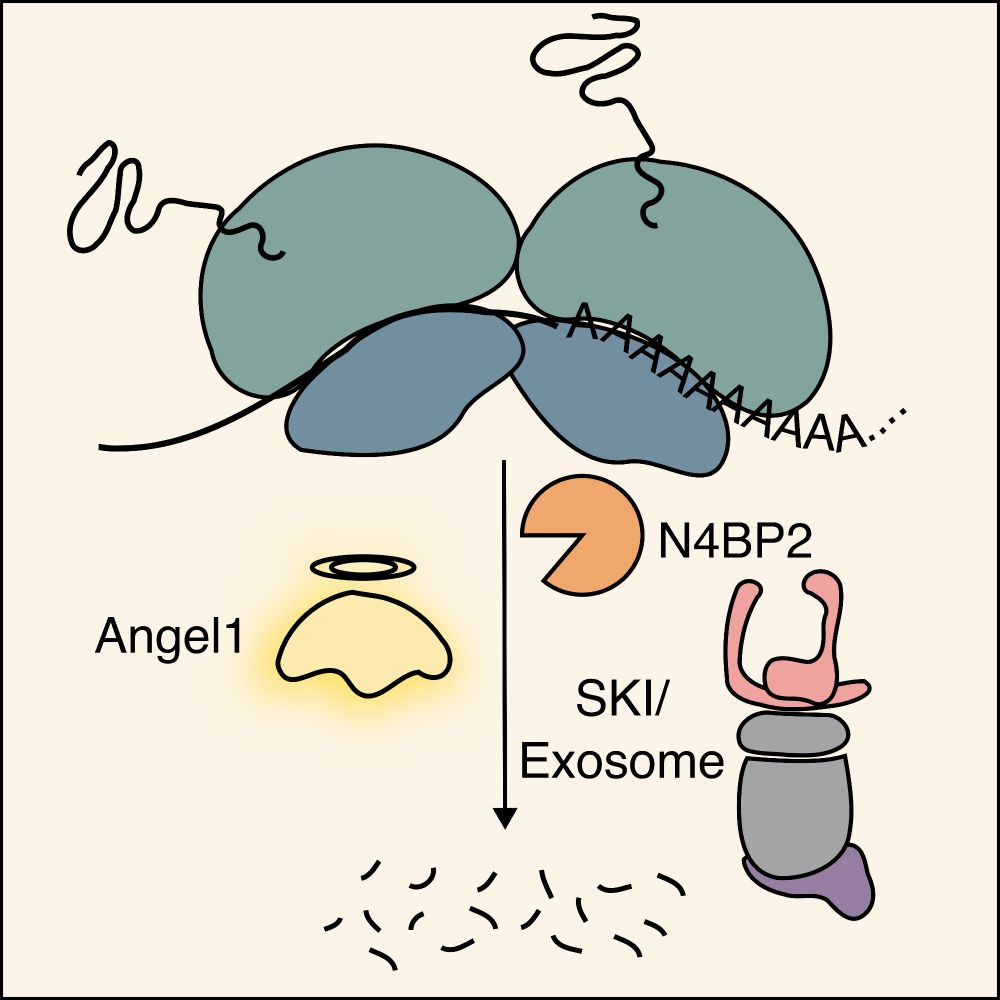
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home

It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social

@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)

We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)
Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology

Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology


Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology
Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology






