
Christine Mordstein
@cmordstein.bsky.social
RNA Therapeutics | Gene Expression Regulation | RNA by Design Leader @ Centillion | Cambridge, UK | Views my own | #RNAsky #RNAtx #RNArocks
Reposted by Christine Mordstein
I highly recommend this meeting! 👌 #RNAsky
Next Codon Usage Meeting, May 31st to June 3rd, 2026 in Montreal right after the RNA Society Meeting. The list of speakers will be up soon.
sites.google.com/view/codonus...
sites.google.com/view/codonus...

codon usage
About the Conference
Codon usage bias—the preference for certain synonymous codons—is a key factor in genome regulation. Codon usage and synonymous codon mutations have been shown to influence gene ex...
sites.google.com
November 7, 2025 at 6:43 PM
I highly recommend this meeting! 👌 #RNAsky
Reposted by Christine Mordstein
Colliding ribosomes are potent signals of cellular stress. But do cells use ‘programmed’ ribosome collisions to regulate gene expression? I’m excited to present a new story from my lab led by Frederick Rehfeld(@fred-rehfeld.bsky.social) which revealed that the answer is YES! Read on to find out how👇

Oxidative stress sensing by the translation elongation machinery promotes production of detoxifying selenoproteins
Selenocysteine, incorporated into polypeptides at recoded termination codons, plays an essential role in redox biology. Using GPX1 and GPX4, selenoenzymes that mitigate oxidative stress, as reporters,...
www.biorxiv.org
October 14, 2025 at 10:28 PM
Colliding ribosomes are potent signals of cellular stress. But do cells use ‘programmed’ ribosome collisions to regulate gene expression? I’m excited to present a new story from my lab led by Frederick Rehfeld(@fred-rehfeld.bsky.social) which revealed that the answer is YES! Read on to find out how👇
Reposted by Christine Mordstein
Two group leader positions available in the broader areas of RNA science, RNA technologies, and RNA medicine. Attractive packages and a great environment. Come and join us at Helmholtz RNA Würzburg, Bavaria.

October 8, 2025 at 9:45 PM
Two group leader positions available in the broader areas of RNA science, RNA technologies, and RNA medicine. Attractive packages and a great environment. Come and join us at Helmholtz RNA Würzburg, Bavaria.
Chemical mods on RNA (whether transient or stable) are the way to go in #RNAtx
New and simple chemical modifications to poly(A) tails extend protein expression -- a really easy one-pot method
Great work by Linglan Fang and Wenrui Zhong in our lab!
doi.org/10.1021/jacs.5c11900
Great work by Linglan Fang and Wenrui Zhong in our lab!
doi.org/10.1021/jacs.5c11900

Localized 2′-OH Acylation at Poly(A) Extends RNA Translation
The potential of coding RNAs as a general therapeutic modality is limited by their short intracellular lifetime. Here, we investigate the effects of localized post-transcriptional RNA modification on ...
doi.org
September 6, 2025 at 11:25 AM
Chemical mods on RNA (whether transient or stable) are the way to go in #RNAtx
Reposted by Christine Mordstein
Application for 2025/2026 RNA Salons is now open. Please apply by 31st August to receive support your local or regional RNA activities. @rnasociety.bsky.social #Lexogen www.rnasociety.org/salons
www.rnasociety.org
August 17, 2025 at 8:32 AM
Application for 2025/2026 RNA Salons is now open. Please apply by 31st August to receive support your local or regional RNA activities. @rnasociety.bsky.social #Lexogen www.rnasociety.org/salons
Reposted by Christine Mordstein
This preprint from Helen Sakharova is one of the coolest things to come out of my lab: “Protein language models reveal evolutionary constraints on synonymous codon choice.” Codon choice is a big puzzle in how information is encoded in genomes, and we have a new angle. www.biorxiv.org/content/10.1...

Protein language models reveal evolutionary constraints on synonymous codon choice
Evolution has shaped the genetic code, with subtle pressures leading to preferences for some synonymous codons over others. Codons are translated at different speeds by the ribosome, imposing constrai...
www.biorxiv.org
August 7, 2025 at 8:29 AM
This preprint from Helen Sakharova is one of the coolest things to come out of my lab: “Protein language models reveal evolutionary constraints on synonymous codon choice.” Codon choice is a big puzzle in how information is encoded in genomes, and we have a new angle. www.biorxiv.org/content/10.1...
My search for a high range #RNA ladder (up to ~11kb) is turning out more tricky than anticipated. The only one I could find is from a supplier that is not too straight forward to order from. Any pointers to suitable products that I may have missed would be greatly appreciated! #RNAsky #RNAtx
July 30, 2025 at 8:25 AM
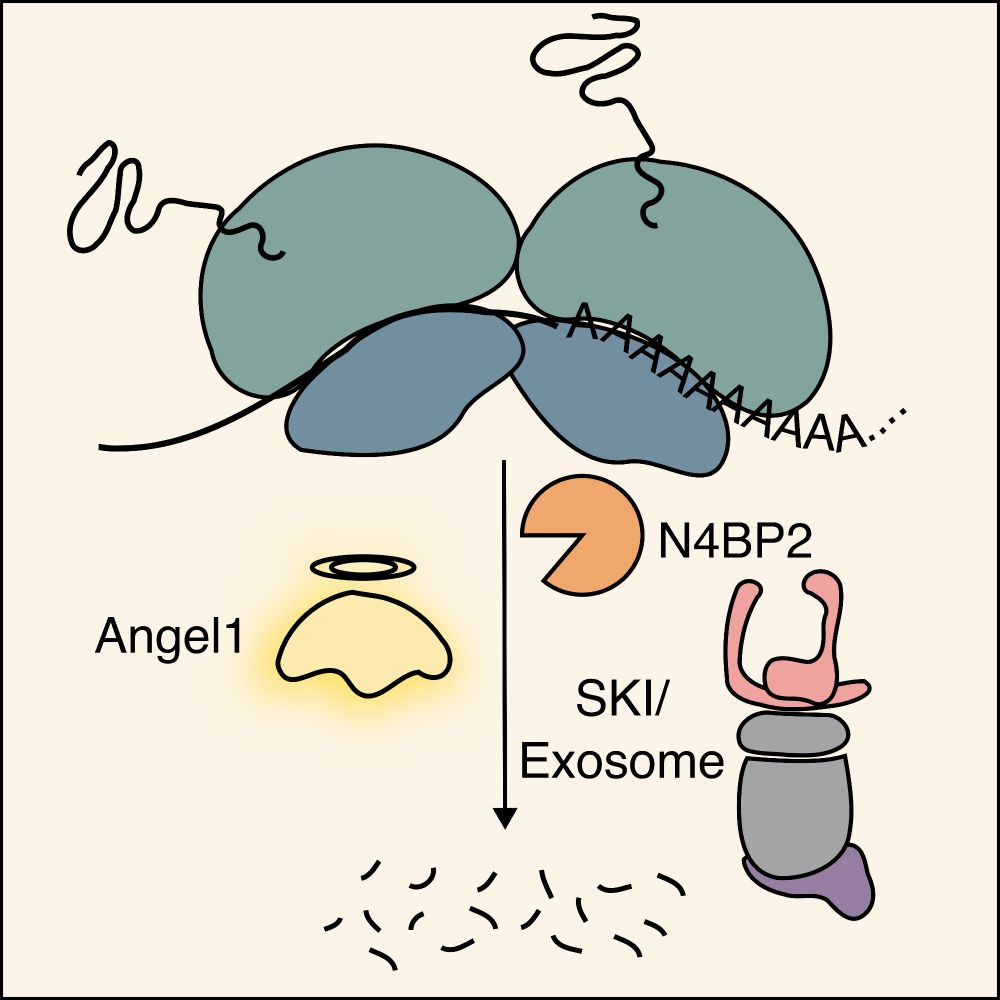
Human CCR4 deadenylase homolog Angel1 along with endonuclease N4BP2 and the SKI-exosome complex promotes degradation of aberrant mRNAs that lack stop codons causing ribosome stalling at the poly(A)-tail bit.ly/4lsYWC6
July 23, 2025 at 5:05 PM
Reposted by Christine Mordstein
Hi #RNAsky, #ChromatinSky!
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home

Home
rna.web.ox.ac.uk
July 18, 2025 at 9:47 AM
Hi #RNAsky, #ChromatinSky!
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
Reposted by Christine Mordstein
We are seeking speakers for the upcoming academic year! Contact us if you are interested!
@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social
@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social

July 16, 2025 at 9:48 AM
We are seeking speakers for the upcoming academic year! Contact us if you are interested!
@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social
@cambiochem.bsky.social #CambridgeRNAClub #RNAClub @rnasociety.bsky.social
Reposted by Christine Mordstein
🚨 New preprint alert 🚨
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)

Systematic benchmarking of basecalling models for RNA modification detection with highly multiplexed nanopore sequencing
Nanopore direct RNA sequencing (DRS) holds promise for advancing our understanding of the epitranscriptome by detecting RNA modifications in native RNA molecules. Recently, Oxford Nanopore Technologie...
bit.ly
July 14, 2025 at 4:00 PM
🚨 New preprint alert 🚨
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)
Reposted by Christine Mordstein
Love RNA biology and care about reproducibility? I’m co-editing a @bio-protocol.bsky.social special issue with @marionhogg.bsky.social & Renate Weizbauer —focusing open access and transparent RNA detection protocols.
Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology
Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology

July 12, 2025 at 1:34 PM
Love RNA biology and care about reproducibility? I’m co-editing a @bio-protocol.bsky.social special issue with @marionhogg.bsky.social & Renate Weizbauer —focusing open access and transparent RNA detection protocols.
Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology
Submit yours today - en.bio-protocol.org/content17
#OpenScience #ReproRNAbiology
Reposted by Christine Mordstein
Ribosomes have been shown to synthesize cyclic peptides with ring-shaped backbones, offering new possibilities for stable, targeted drug development and advanced biomaterials. doi.org/g9sm8h

First-ever ribosomal synthesis of cyclic peptides opens new avenues for next-generation drug design
Inside our cells, ribosomes—the tireless "protein factories" of life—have just shown off a new skill they haven't used in billions of years.
phys.org
July 8, 2025 at 9:02 PM
Ribosomes have been shown to synthesize cyclic peptides with ring-shaped backbones, offering new possibilities for stable, targeted drug development and advanced biomaterials. doi.org/g9sm8h
Reposted by Christine Mordstein
BREAKING: Scientists are staging a “science fair” in the lobby of a Congressional building to tell elected officials about the critical knowledge the US will lose because their research grants have been canceled.
July 8, 2025 at 3:31 PM
BREAKING: Scientists are staging a “science fair” in the lobby of a Congressional building to tell elected officials about the critical knowledge the US will lose because their research grants have been canceled.
Reposted by Christine Mordstein
UK bosses to be banned from using NDAs to cover up misconduct at work

UK bosses to be banned from using NDAs to cover up misconduct at work
Exclusive: Changes to workers’ rights bill will prohibit the silencing of staff who suffer harassment or discrimination
*
Labour has delivered where Tory ministers did not on NDA rules
Bosses in the UK will be banned from using non-disclosure agreements to silence employees who have suffered harassment and discrimination in the workplace as part of the government’s overhaul of workers’ rights.
Ministers will on Monday night table amendments to the government’s employment rights bill to prohibit the widespread practice of using legally enforceable NDAs to conceal unacceptable behaviour at work. Continue reading...
www.theguardian.com
July 7, 2025 at 6:11 PM
UK bosses to be banned from using NDAs to cover up misconduct at work
En route to #Edinburgh for the week! 🏴
Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology
Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology
July 6, 2025 at 10:27 AM
En route to #Edinburgh for the week! 🏴
Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology
Looking forward to catching up with friends and former colleagues at #MRC_HGU #IGC and across #EdinburghUni 😀 If you are based in Edinburgh and interested in tapping into the #RNAtx space, give me shout! #RNAsky #RNAbiology
Any recommendations for outsourcing Sanger sequencing? Looking for a reliable service in the UK with a short turnaround time. Bonus points if it's in Cambridge 😉 #MolecularBiology #RNAsky #oldschool 🧪
July 5, 2025 at 5:24 PM
Any recommendations for outsourcing Sanger sequencing? Looking for a reliable service in the UK with a short turnaround time. Bonus points if it's in Cambridge 😉 #MolecularBiology #RNAsky #oldschool 🧪
Reposted by Christine Mordstein
Thank you! Yes, absolutely! We wouldn't know what to do without it. I strongly recommend people check it out! www.rnacanvas.app
RNAcanvas
A web app for drawing and exploring nucleic acid structures.
www.rnacanvas.app
July 2, 2025 at 6:58 PM
Thank you! Yes, absolutely! We wouldn't know what to do without it. I strongly recommend people check it out! www.rnacanvas.app
Reposted by Christine Mordstein
WarpDemuX boosts nanopore direct RNA sequencing with 2x faster and more accurate demultiplexing using a lightweight ML algorithm for SQK-RNA002/004 kits. PMID:40258808, Nat Commun 2025, @NatureComms https://doi.org/10.1038/s41467-025-59102-9 #Medsky #Pharmsky #RNA #ASHG #ESHG 🧪

Demultiplexing and barcode-specific adaptive sampling for nanopore direct RNA sequencing | Nature Communications
Nanopore direct RNA sequencing (dRNA-seq) enables unique insights into RNA biology. However, applications are currently limited by the lack of accurate and cost-effective sample multiplexing. Here we introduce WarpDemuX, an ultra-fast and highly accurate adapter-barcoding and demultiplexing approach for dRNA-seq with SQK-RNA002 and SQK-RNA004 chemistries. WarpDemuX enhances speed and accuracy by fast processing of the raw nanopore signal, use of a light-weight machine-learning algorithm and design of optimized barcode sets. We demonstrate its utility by performing rapid phenotypic profiling of different SARS-CoV-2 viruses through multiplexed sequencing of longitudinal samples on a single flowcell, identifying systematic differences in transcript abundance and poly(A) tail lengths during infection. Additionally, integrating WarpDemuX into sequencing control software enables real-time enrichment of target molecules through barcode-specific adaptive sampling, which we demonstrate by enric
doi.org
June 23, 2025 at 5:10 AM
Reposted by Christine Mordstein
Mechanism of release factor–mediated peptidyl-tRNA hydrolysis on the ribosome www.science.org/doi/10.1126/...

Mechanism of release factor–mediated peptidyl-tRNA hydrolysis on the ribosome
Translation termination is essential in all living organisms because it ensures that proteins have lengths strictly defined by their genes. This universal process is mediated by peptide release factor...
www.science.org
June 19, 2025 at 7:39 PM
Mechanism of release factor–mediated peptidyl-tRNA hydrolysis on the ribosome www.science.org/doi/10.1126/...
Reposted by Christine Mordstein
Pan-viral ORFs discovery using massively parallel ribosome profiling www.science.org/doi/10.1126/...

Pan-viral ORFs discovery using massively parallel ribosome profiling
Defining viral proteomes is crucial to understanding viral life cycles and immune recognition but the landscape of translated regions remains unknown for most viruses. We have developed massively para...
www.science.org
June 20, 2025 at 6:36 PM
Pan-viral ORFs discovery using massively parallel ribosome profiling www.science.org/doi/10.1126/...
Reposted by Christine Mordstein
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
June 13, 2025 at 4:30 AM
EPIGENETIC HULK READY TO SMASH AGAIN! GET IN LOSERS!
Reposted by Christine Mordstein
Cytoplasmic DIS3 is an exosome-independent endoribonuclease with catalytic activity toward circular RNAs
Cytoplasmic DIS3 is an exosome-independent endoribonuclease with catalytic activity toward circular RNAs
CircRNAs are generated during splicing, but their specific degradation pathways are poorly understood. Latini et al. identify and biochemically characterize the exo- and endoribonuclease DIS3 as a candidate for cytoplasmic circRNA degradation. DIS3 uses its catalytic PIN domain to cleave circRNAs and functions in the cytoplasm independently of the exosome.
dlvr.it
June 1, 2025 at 2:38 PM
Cytoplasmic DIS3 is an exosome-independent endoribonuclease with catalytic activity toward circular RNAs



