
I'm Jay. I study the ecology and evolution of plant-insect interactions using a combination of omics approaches and field experimentation. I'm especially interested in how toxic plants in the nightshade family have co-evolved with specialist herbivores.
Wednesday Dec 10th at 9am PT I'll livestream an interactive demo of what I have learned (matsen.group/agentic.html) about how to leverage agentic coding to do rigorous science.
www.youtube.com/watch?v=Rbhs...

🌴 🐸 🐒 🦎 🌱
@morphoinstitute.org
July 11-25, 2026 at the Amazon Conservatory for Tropical Studies (morphoinstitute.org/actsperu/)
For more info and to apply: forms.gle/hxgaiPuYhdfD...

🌴 🐸 🐒 🦎 🌱
@morphoinstitute.org
July 11-25, 2026 at the Amazon Conservatory for Tropical Studies (morphoinstitute.org/actsperu/)
For more info and to apply: forms.gle/hxgaiPuYhdfD...
Thanks to @bhuwanabbot.bsky.social
for leading and collaborators:
@raw937.bsky.social @andrabuchan.bsky.social
academic.oup.com/gbe/advance-...

Thanks to @bhuwanabbot.bsky.social
for leading and collaborators:
@raw937.bsky.social @andrabuchan.bsky.social
academic.oup.com/gbe/advance-...
1/n
Them: *sends blurry photo of a speck on a wall* "hey do you know what this bug in my house is?"
Them: *sends blurry photo of a speck on a wall* "hey do you know what this bug in my house is?"
www.cell.com/trends/genet...

www.cell.com/trends/genet...
🔗 www.biorxiv.org/content/10.1...

🔗 www.biorxiv.org/content/10.1...


www.cell.com/trends/ecolo...

www.cell.com/trends/ecolo...
Except weevils. It's pretty bad at annotating my weevil genomes.
No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

Except weevils. It's pretty bad at annotating my weevil genomes.
We find the fly development gene bicoid is much older than previously thought (~20 million yrs older!) 🪰🧬
To pinpoint its origins we tackled the Diptera phylogeny, providing some resolution (many open questions remain).
🔗 tinyurl.com/2vyuevpy
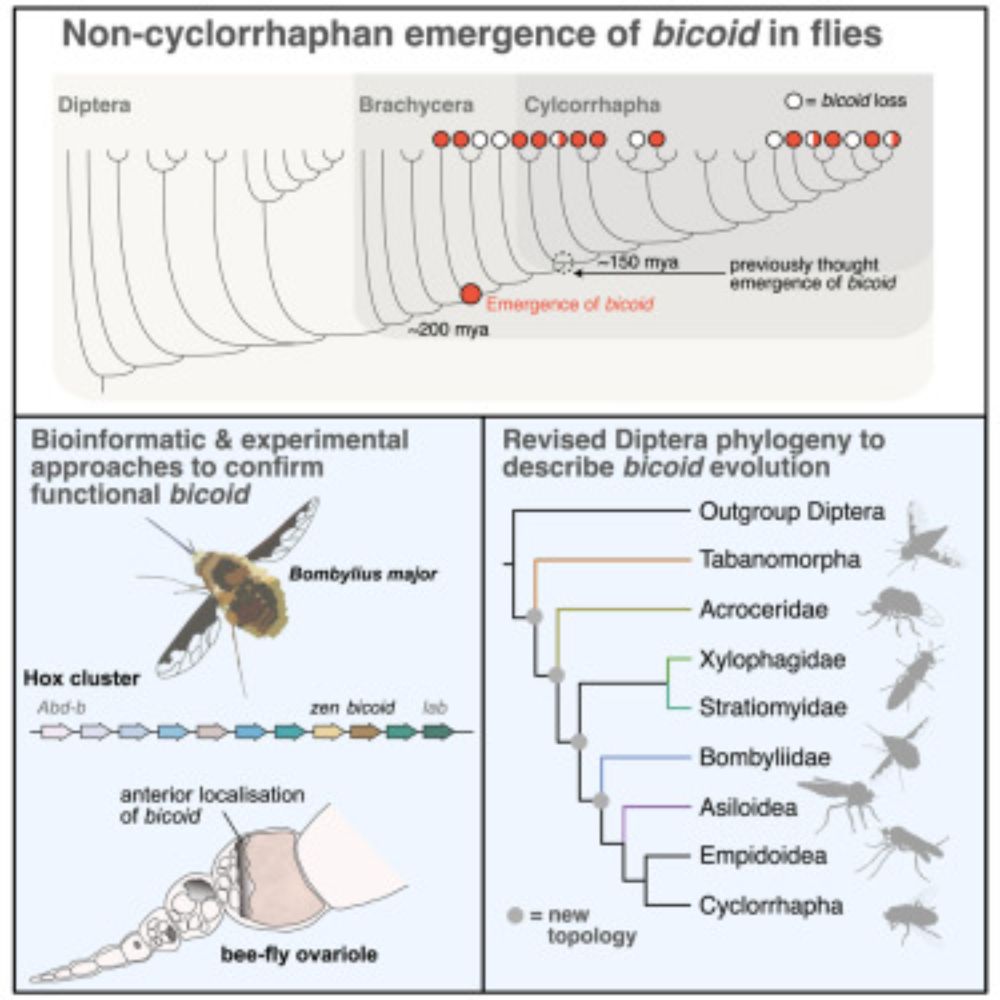
We find the fly development gene bicoid is much older than previously thought (~20 million yrs older!) 🪰🧬
To pinpoint its origins we tackled the Diptera phylogeny, providing some resolution (many open questions remain).
🔗 tinyurl.com/2vyuevpy
Using long-term Åland monitoring data, we found that herbivory increases plant diversity across scales and flips the diversity–area relationship: a positive relationship is found in grazed sites while a negative one in ungrazed sites.
🔗 doi.org/10.1111/ele....

Using long-term Åland monitoring data, we found that herbivory increases plant diversity across scales and flips the diversity–area relationship: a positive relationship is found in grazed sites while a negative one in ungrazed sites.
🔗 doi.org/10.1111/ele....

Investigating the evolution of large meiotic rings of multiple X and Y chromosomes in two Leptodactylus species
🔗 www.nature.com/articles/s42...

Investigating the evolution of large meiotic rings of multiple X and Y chromosomes in two Leptodactylus species
🔗 www.nature.com/articles/s42...
apply.interfolio.com/176994
apply.interfolio.com/176994
Apps reviewed 12/15.
Please spread the word! 💚⛏️
#PlantSci 🧪🦠🧬
jobs.charlotte.edu/postings/65141
Apps reviewed 12/15.
Please spread the word! 💚⛏️
#PlantSci 🧪🦠🧬
jobs.charlotte.edu/postings/65141
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics
pubmed.ncbi.nlm.nih.gov/24378633/


