
The 2nd Nanopore Users Day will take place 🗓️10 March 2026 at the Duclaux Amphitheater, Institut Pasteur (Paris). This edition will be open to external participants!
👉 More details and registration info coming soon!
@chloebaum.bsky.social @pasteur.fr @nanoporetech.com

The 2nd Nanopore Users Day will take place 🗓️10 March 2026 at the Duclaux Amphitheater, Institut Pasteur (Paris). This edition will be open to external participants!
👉 More details and registration info coming soon!
@chloebaum.bsky.social @pasteur.fr @nanoporetech.com

Join our live webinar tomorrow. https://bit.ly/47RharD

Join our live webinar tomorrow. https://bit.ly/47RharD
Register: nanoporetech.swoogo.com/Nanopore_X_L...
Register: nanoporetech.swoogo.com/Nanopore_X_L...
With @rdrpsummit.bsky.social - excited to announce talk and discussion with Dr Sanguu Kim about HF nanopore sequencing strategy for virology :)
calendar.app.google/oyRVP6v5Tvjg...
CET: 14:00–15:00,
Format: 30 min + 30 min

With @rdrpsummit.bsky.social - excited to announce talk and discussion with Dr Sanguu Kim about HF nanopore sequencing strategy for virology :)
calendar.app.google/oyRVP6v5Tvjg...
CET: 14:00–15:00,
Format: 30 min + 30 min
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
doi.org/10.1101/2025...

doi.org/10.1101/2025...
A powerful new tool for real-time analysis of #nanopore #metagenomics 👇

A powerful new tool for real-time analysis of #nanopore #metagenomics 👇


doi.org/10.1371/jour...

doi.org/10.1371/jour...
📑Fine Mapping Regulatory Variants by Characterizing Native CpG Methylation with Nanopore Long-Read Sequencing
👉 bit.ly/4o8qMoY

📑Fine Mapping Regulatory Variants by Characterizing Native CpG Methylation with Nanopore Long-Read Sequencing
👉 bit.ly/4o8qMoY
"Nanopore long-read sequencing for the critically ill facilitates ultrarapid diagnostics and urgent clinical decision making"
#raredisease #WGS @nanoporetech.com @erasmusmc.bsky.social
www.nature.com/articles/s41...

"Nanopore long-read sequencing for the critically ill facilitates ultrarapid diagnostics and urgent clinical decision making"
#raredisease #WGS @nanoporetech.com @erasmusmc.bsky.social
www.nature.com/articles/s41...
Learn how to sequence full length 16S and ITS genes with Oxford Nanopore's latest kit. Recordings also sent out after signup.
nanoporetech.com/about/events... #16S #ITS #metagenomics #metabarcoding

Learn how to sequence full length 16S and ITS genes with Oxford Nanopore's latest kit. Recordings also sent out after signup.
nanoporetech.com/about/events... #16S #ITS #metagenomics #metabarcoding
- Plasmid EZ: Easy as 1, 2, 3
- Nanopore Technology Applications at Azenta Life Sciences
- Benefits of mixed platform multiomics
#BiopharmaDay #Nanopore #NGS #ONT



- Plasmid EZ: Easy as 1, 2, 3
- Nanopore Technology Applications at Azenta Life Sciences
- Benefits of mixed platform multiomics
#BiopharmaDay #Nanopore #NGS #ONT
www.biorxiv.org/content/10.1...
#AMR #Nanopore #PublicHealth #GenomicSurveillance

www.biorxiv.org/content/10.1...
#AMR #Nanopore #PublicHealth #GenomicSurveillance
📺 You can watch here: www.youtube.com/watch?v=t5cQ...
#LongReads #AMR #antimicrobial #metagenomics #microbiome #nanopore
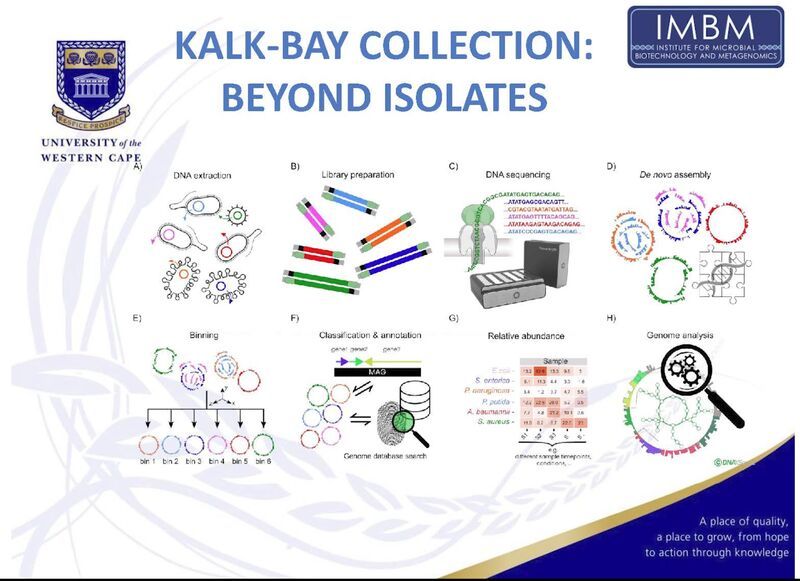
📺 You can watch here: www.youtube.com/watch?v=t5cQ...
#LongReads #AMR #antimicrobial #metagenomics #microbiome #nanopore

biorxiv.org/content/10.1101/2025.09.23.678181v1
#NanoporeSequencing #Genomics #TB

biorxiv.org/content/10.1101/2025.09.23.678181v1
#NanoporeSequencing #Genomics #TB

