Runner.
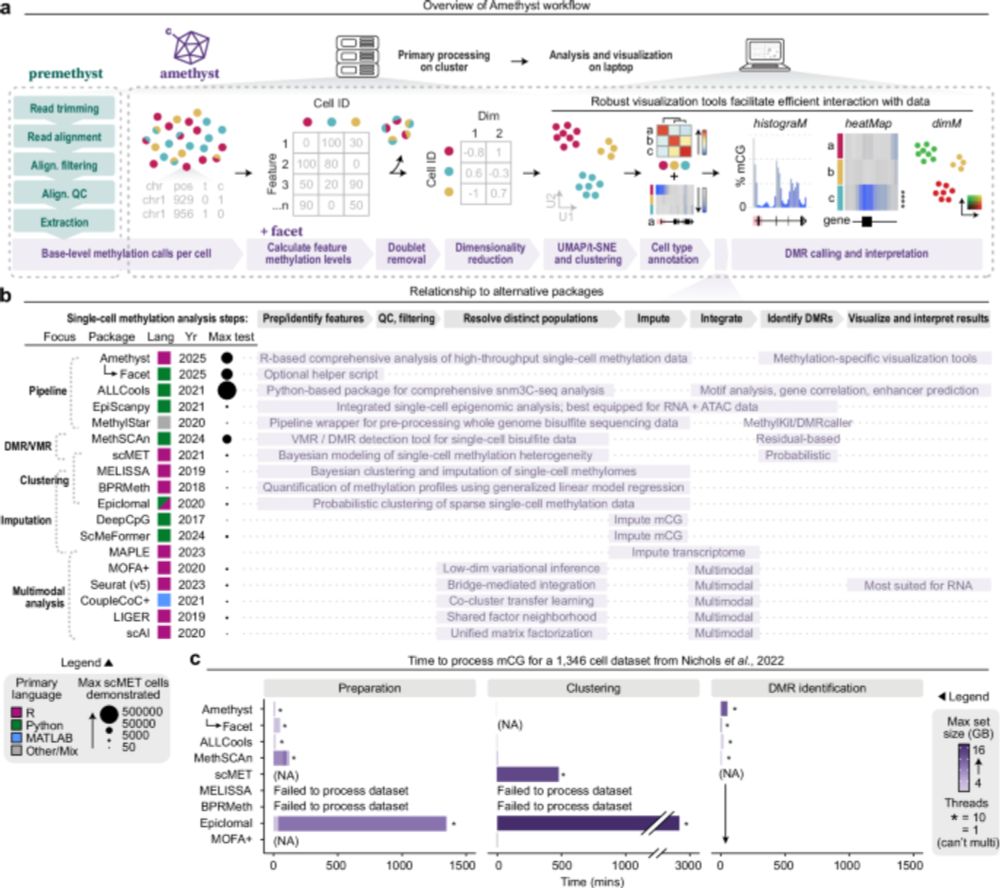
www.nature.com/articles/s42...
www.nature.com/articles/s42...

www.nature.com/articles/s42...
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
@jbuenrostro.bsky.social has been named core institute member of the Broad. His lab will study how cells change in response to life experiences, and how those alterations affect health and disease.


Learn more: bit.ly/4mAWAS8

github.com/vierstralab/...
github.com/vierstralab/...
"Molecular circuits for genomic recording of cellular events"
by Wei Chen & Junhong Choi (@choijunhong.bsky.social)
FREE to read till June 25th using this link:
authors.elsevier.com/a/1l2gMcQbJF...



www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics

www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
Also, I know this is extremely unnerving if u weren't expecting this. But let's not panic. Let's organize & give this the best fking fight of our lives.
Also, I know this is extremely unnerving if u weren't expecting this. But let's not panic. Let's organize & give this the best fking fight of our lives.
(Plus lots more)
www.cell.com/cell-genomic...

(Plus lots more)
www.cell.com/cell-genomic...

