
We’re excited to welcome @landthalerm.bsky.social for the upcoming #CRC1678 Seminar!
🧬 He’ll dive into: "Posttranscriptional Regulation in Cellular Time and Space"
📅 November 25
📍 Auditorium, MPI-Age
⏰ 2:00 PM
Explore how RNA dynamics shape cellular behavior across time and space!
We’re excited to welcome @landthalerm.bsky.social for the upcoming #CRC1678 Seminar!
🧬 He’ll dive into: "Posttranscriptional Regulation in Cellular Time and Space"
📅 November 25
📍 Auditorium, MPI-Age
⏰ 2:00 PM
Explore how RNA dynamics shape cellular behavior across time and space!
nccr-rna-and-disease.ch/news/article...
@omuhlemann-lab.bsky.social
@unibe.ch
www.cell.com/issue/S2666-...
www.cell.com/issue/S2666-...
We report variable activity of the NMD pathway (mRNA QC) across tissues & individuals:
🧠less active in the nervous system & in reproductive tissues
🩸more active in digestive tract, muscle, blood

We report variable activity of the NMD pathway (mRNA QC) across tissues & individuals:
🧠less active in the nervous system & in reproductive tissues
🩸more active in digestive tract, muscle, blood
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
the correct link to the collaboration with Torben and Clemens is:
www.biorxiv.org/content/10.1...
the correct link to the collaboration with Torben and Clemens is:
www.biorxiv.org/content/10.1...
The database ➡️ nmdrht.uni-koeln.de
The database ➡️ nmdrht.uni-koeln.de
Researchers from the #unicologne and the @mdc-berlin.bsky.social have cracked the code of a crucial mechanism of cellular quality control in humans. 👇
uni.koeln/RRCLZ


The University Council has elected Professor Niessen as Vice-Rector for Research to support the advancement of excellent research at the #UniCologne. 🎓
Read more 👇
uni.koeln/9FCEC
@cecad.bsky.social

The University Council has elected Professor Niessen as Vice-Rector for Research to support the advancement of excellent research at the #UniCologne. 🎓
Read more 👇
uni.koeln/9FCEC
@cecad.bsky.social

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
by Kathi Zarnack (@zarnack-group.bsky.social) & colleagues
"This review explores [m6A in] mRNA decay, with a focus on the newly discovered CDS–m6A decay (CMD) pathway..."
Read it here:
authors.elsevier.com/sd/article/S...
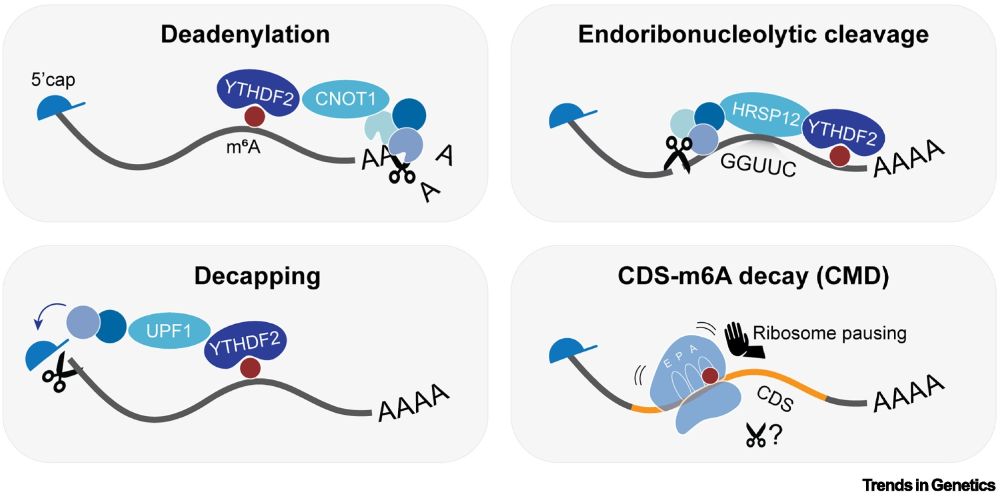
by Kathi Zarnack (@zarnack-group.bsky.social) & colleagues
"This review explores [m6A in] mRNA decay, with a focus on the newly discovered CDS–m6A decay (CMD) pathway..."
Read it here:
authors.elsevier.com/sd/article/S...

www.nature.com/articles/s41...

www.nature.com/articles/s41...

mbg.au.dk/en/news-and-...

mbg.au.dk/en/news-and-...
Please RT!
We're looking for a postdoc to join an exciting joint project between our lab @upf.edu & @crg.eu (Barcelona) and the Sander lab @mdc-berlin.bsky.social (Berlin) investigating how alternative splicing and microexons influences the maturation of pancreatic islets.
Deadline: 30/09/25👇
Please RT!
We're looking for a postdoc to join an exciting joint project between our lab @upf.edu & @crg.eu (Barcelona) and the Sander lab @mdc-berlin.bsky.social (Berlin) investigating how alternative splicing and microexons influences the maturation of pancreatic islets.
Deadline: 30/09/25👇
It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home

It is a pleasure to announce that the @ox.ac.uk RNA & transcription club is now on Bluesky. Follow us to stay in touch with everything RNA and transcription from gene regulation to disease. Join us here: rna.web.ox.ac.uk/home
www.nature.com/articles/s41... Intracellular pathogen effector reprograms host gene expression by inhibiting mRNA decay | Nature Communications
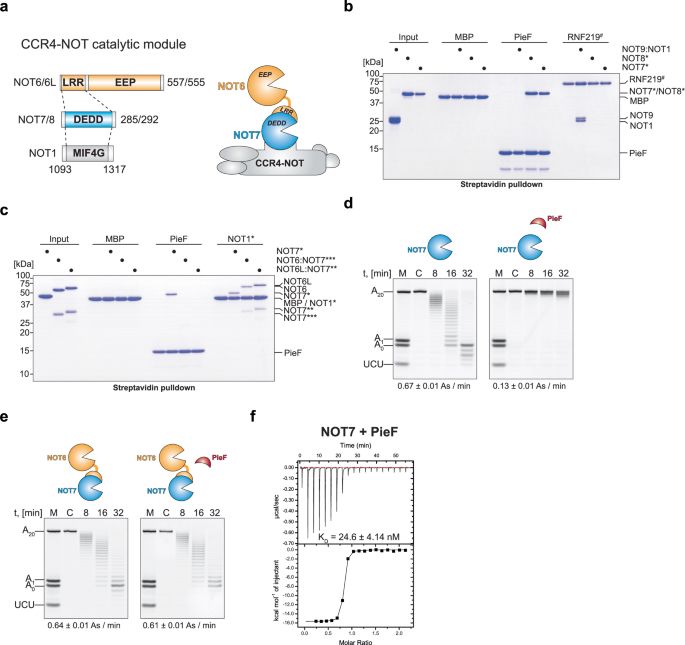
www.nature.com/articles/s41... Intracellular pathogen effector reprograms host gene expression by inhibiting mRNA decay | Nature Communications
👉 crc1678.uni-koeln.de
👉 crc1678.uni-koeln.de


