- IRMPD is the new UPVD
- IRMPD with TMT boosts reporter intensity by up to 240% without affecting quant
- New resolutions -> Down to 1k res, with an associated loss of signal, but at least it looks faster!
- Improved proton transfer reaction
- IRMPD is the new UPVD
- IRMPD with TMT boosts reporter intensity by up to 240% without affecting quant
- New resolutions -> Down to 1k res, with an associated loss of signal, but at least it looks faster!
- Improved proton transfer reaction

@jacs.acspublications.org @ucsfcancer.bsky.social @jimwellsucsf.bsky.social
pubs.acs.org/doi/10.1021/...

@jacs.acspublications.org @ucsfcancer.bsky.social @jimwellsucsf.bsky.social
pubs.acs.org/doi/10.1021/...
The manuscript is available here: www.mcponline.org/article/S153...
The manuscript is available here: www.mcponline.org/article/S153...
#Alzheimers #Glia #Tau #Amyloid

#Alzheimers #Glia #Tau #Amyloid
A cytokine receptor-targeting chimera (kineTAC) toolbox for expanding extracellular targeted protein degradation | bioRxiv
www.biorxiv.org/content/10.1...

A cytokine receptor-targeting chimera (kineTAC) toolbox for expanding extracellular targeted protein degradation | bioRxiv
www.biorxiv.org/content/10.1...
"It’s people who will get cancer in 10, 20, or 30 years who will really pay the price for these cuts."
www.nytimes.com/2025/08/24/o...

"It’s people who will get cancer in 10, 20, or 30 years who will really pay the price for these cuts."
www.nytimes.com/2025/08/24/o...
New reactions are typically developed by trial and error. How can we speed up this process? Read on to learn how we used DNA scaffolding to perform >500,000 parallel reactions on attomole scale.
1/n
New reactions are typically developed by trial and error. How can we speed up this process? Read on to learn how we used DNA scaffolding to perform >500,000 parallel reactions on attomole scale.
1/n
Thank you to Phil and Penny Knight for their incredible generosity.
#GiveCancerHell


Thank you to Phil and Penny Knight for their incredible generosity.
#GiveCancerHell


Excited to continue exploring.
#NIH #NCI #F32 #postdoc #cancerresearch

Excited to continue exploring.
#NIH #NCI #F32 #postdoc #cancerresearch



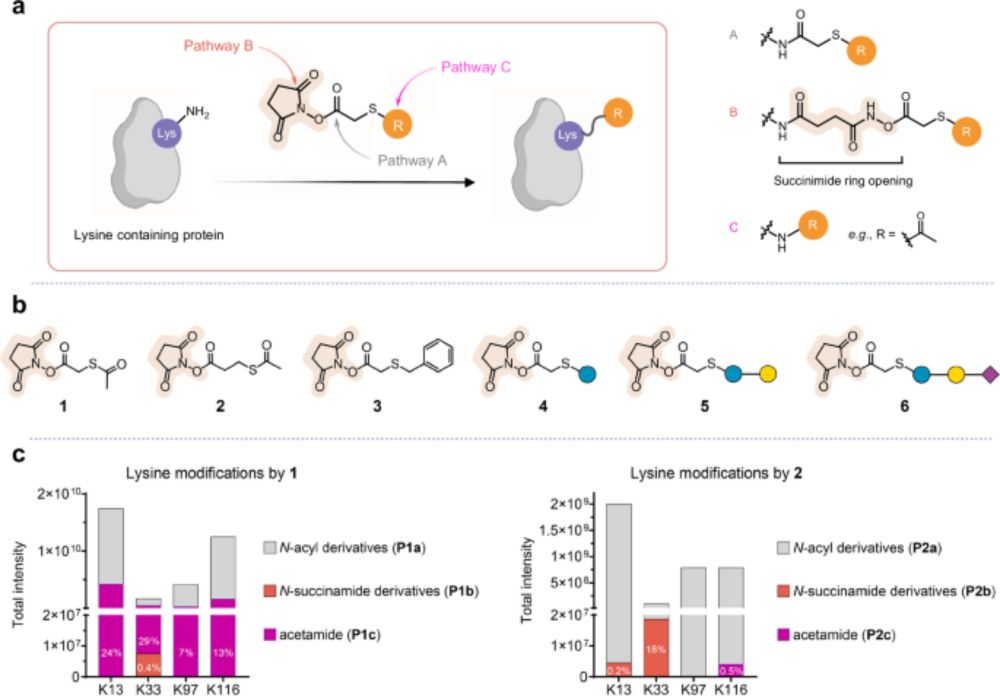
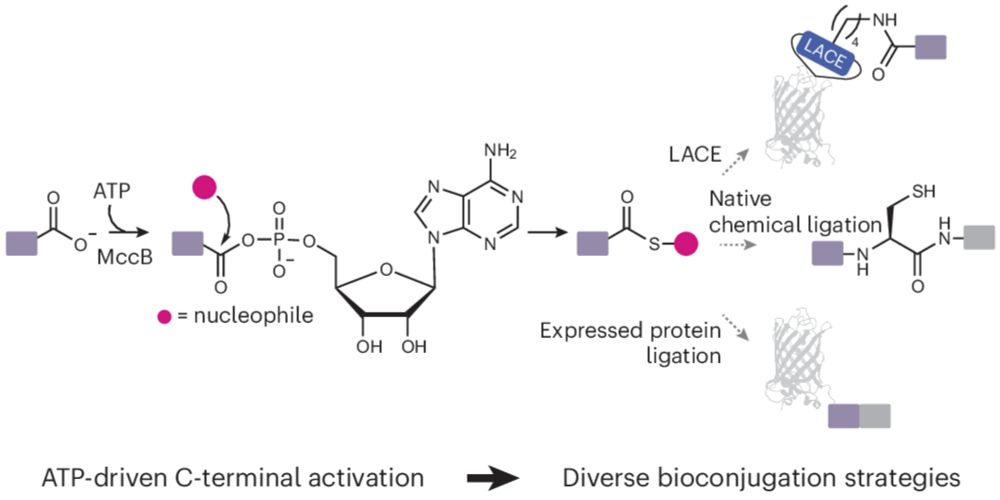
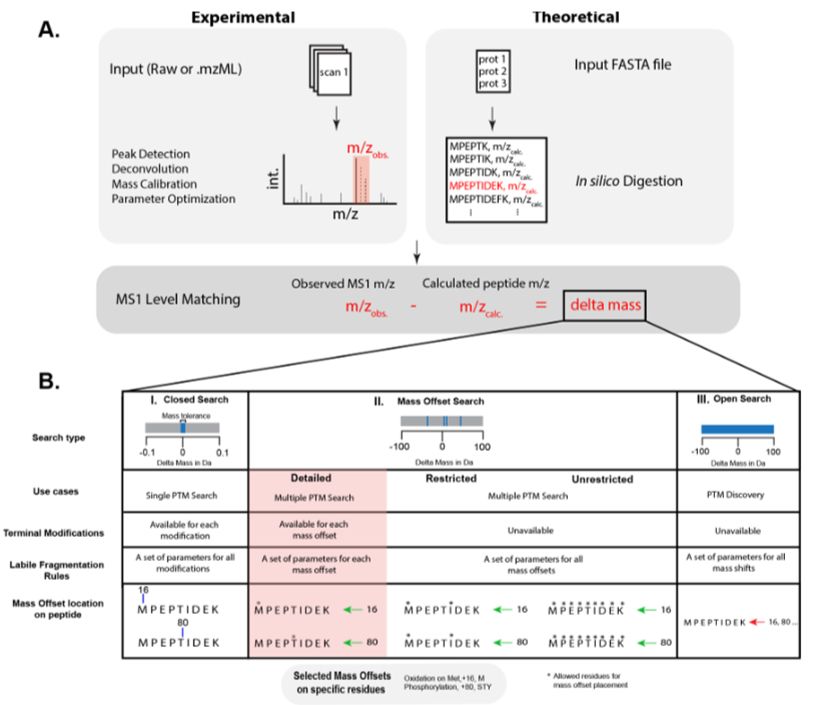
www.nature.com/articles/s41...
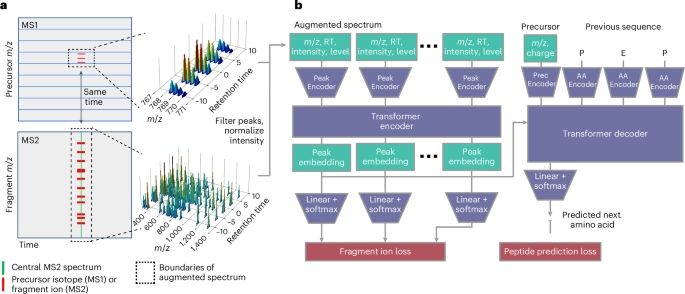
www.nature.com/articles/s41...
Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!



Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!
nationalmaglab.org/careers/meet...

nationalmaglab.org/careers/meet...
www.biorxiv.org/content/10.1...
@ucsfcancer.bsky.social @ucsfhealth.bsky.social @ucberkeleyofficial.bsky.social

www.biorxiv.org/content/10.1...
@ucsfcancer.bsky.social @ucsfhealth.bsky.social @ucberkeleyofficial.bsky.social
I feel so fortunate for this mass spec community as we champion open and accessible science.




I feel so fortunate for this mass spec community as we champion open and accessible science.
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀
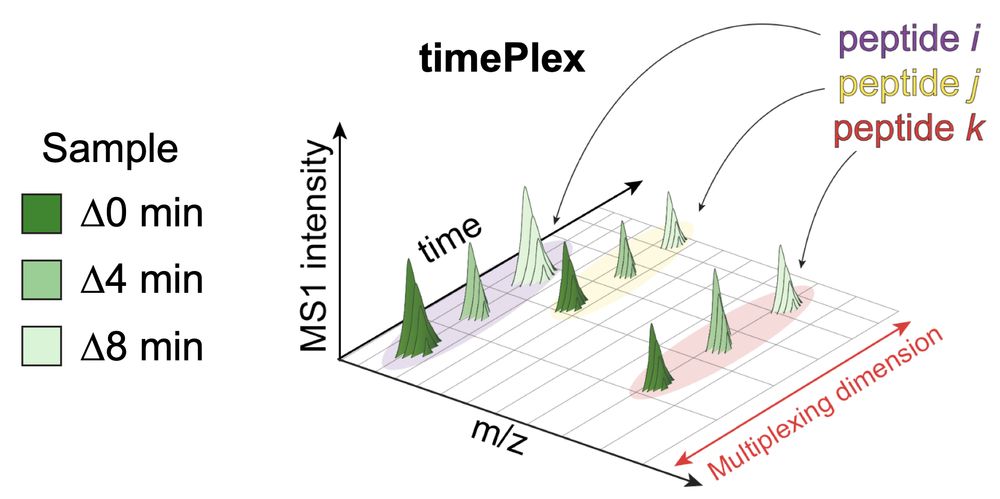
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀
We reviewed modern MS instrument platforms and the acquisition strategies they enable.
Now available on #ChemRxiv: doi.org/10.26434/che...
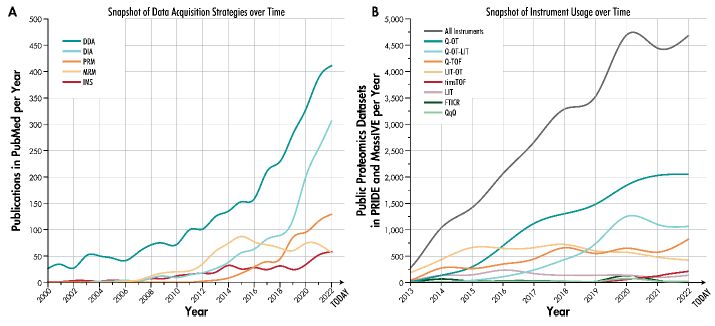
We reviewed modern MS instrument platforms and the acquisition strategies they enable.
Now available on #ChemRxiv: doi.org/10.26434/che...