Past: UW 🦡, Oxford 🇬🇧, NIH 🇺🇸, Toronto 🇨🇦



Great project with a great PI.
A no-miss opportunity
More information :
www.cbs.cnrs.fr/images/jobs/...
The position is expected to start in early 2026, with some flexibility.

Great project with a great PI.
A no-miss opportunity
#ERCSyG #biotechnology
@erc.europa.eu
📷A. Heddergott

#ERCSyG #biotechnology
@erc.europa.eu
📷A. Heddergott
Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity

Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity

Submit an abstract today buff.ly/oOjstk6

Submit an abstract today buff.ly/oOjstk6
Instruct-ERIC provides open access to advanced structural biology technologies. #BNMRZ advances innovation in #NMR methods, drug discovery, and imaging - boosting the German Centre’s capabilities.
👉 Read more: t1p.de/pjtyv

Instruct-ERIC provides open access to advanced structural biology technologies. #BNMRZ advances innovation in #NMR methods, drug discovery, and imaging - boosting the German Centre’s capabilities.
👉 Read more: t1p.de/pjtyv

Join the 10th European Solid-State NMR Summer School❗️
🗓️ October 20-24, 2025 | Garching, Germany
📌 Application Deadline: 01.09.2025
www.bio.nat.tum.de/ocb/upcoming...
#NMRchat #BioNMR
Join the 10th European Solid-State NMR Summer School❗️
🗓️ October 20-24, 2025 | Garching, Germany
📌 Application Deadline: 01.09.2025
www.bio.nat.tum.de/ocb/upcoming...
#NMRchat #BioNMR

I'm thrilled to share our new paper (Adriaenssens et al., Nat Cell Biol 2025).
This paper describes a new mechanism for the initiation of autophagosome biogenesis.
We found that this WIPI-ATG13-driven pathway is preferentially used by a group of transmembrane autophagy receptors.
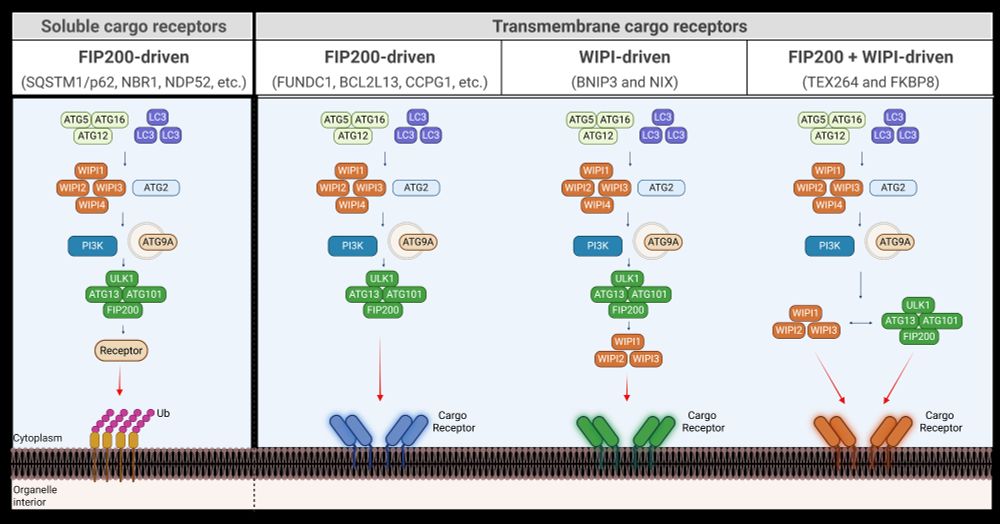
I'm thrilled to share our new paper (Adriaenssens et al., Nat Cell Biol 2025).
This paper describes a new mechanism for the initiation of autophagosome biogenesis.
We found that this WIPI-ATG13-driven pathway is preferentially used by a group of transmembrane autophagy receptors.
For more detailed information, please follow the link:
www.bio.nat.tum.de/ocb/upcoming...
For more detailed information, please follow the link:
www.bio.nat.tum.de/ocb/upcoming...



levitate.bio/alphafold/mu...
doi.org/10.1093/nar/...
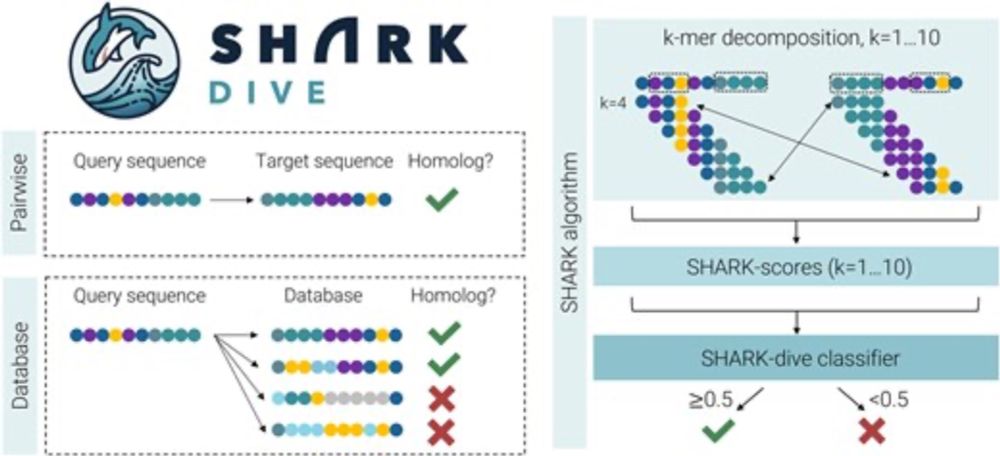
doi.org/10.1093/nar/...
www.nature.com/articles/s41...
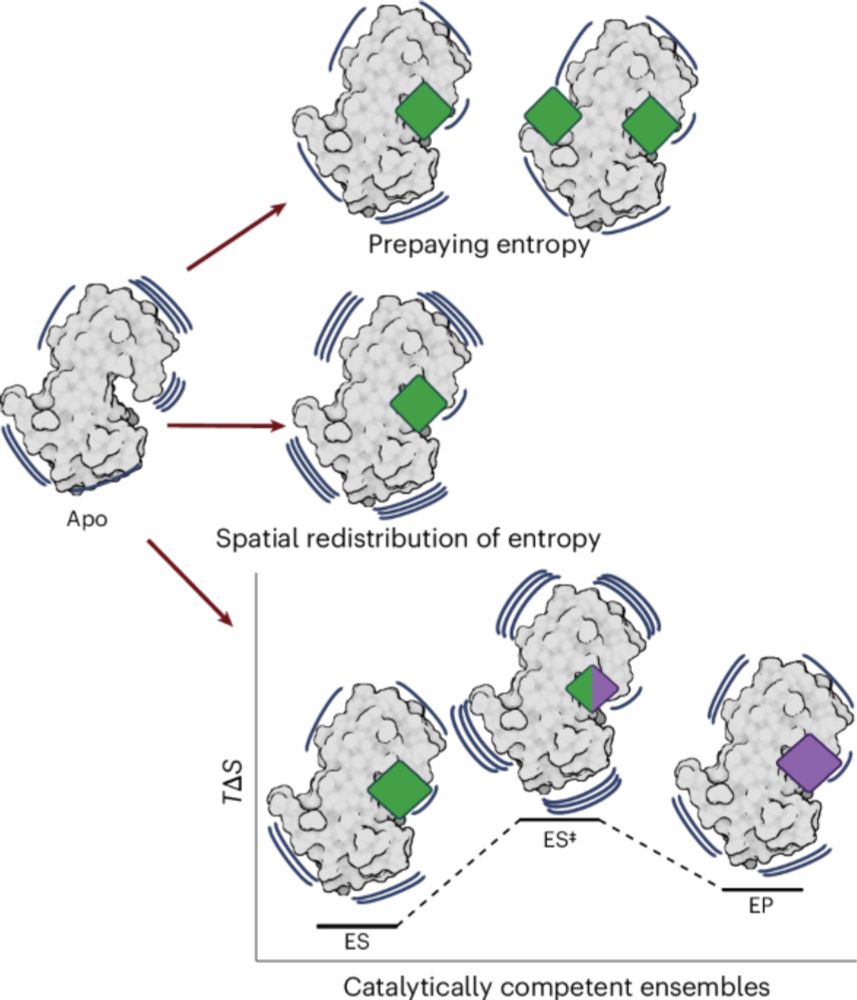
www.nature.com/articles/s41...

jobs.helmholtz-muenchen.de/jobposting/b...
#science #PhD #research

jobs.helmholtz-muenchen.de/jobposting/b...
#science #PhD #research
3 year post. Ad closes 12 March: www.kcl.ac.uk/jobs/105866-...

3 year post. Ad closes 12 March: www.kcl.ac.uk/jobs/105866-...