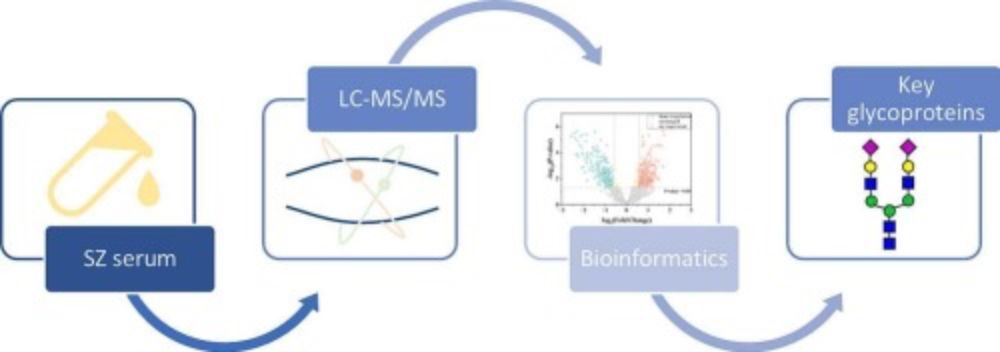
onlinelibrary.wiley.com/doi/full/10.... #Bioinformatics #microbiome #metaproteomics #proteomics #MS
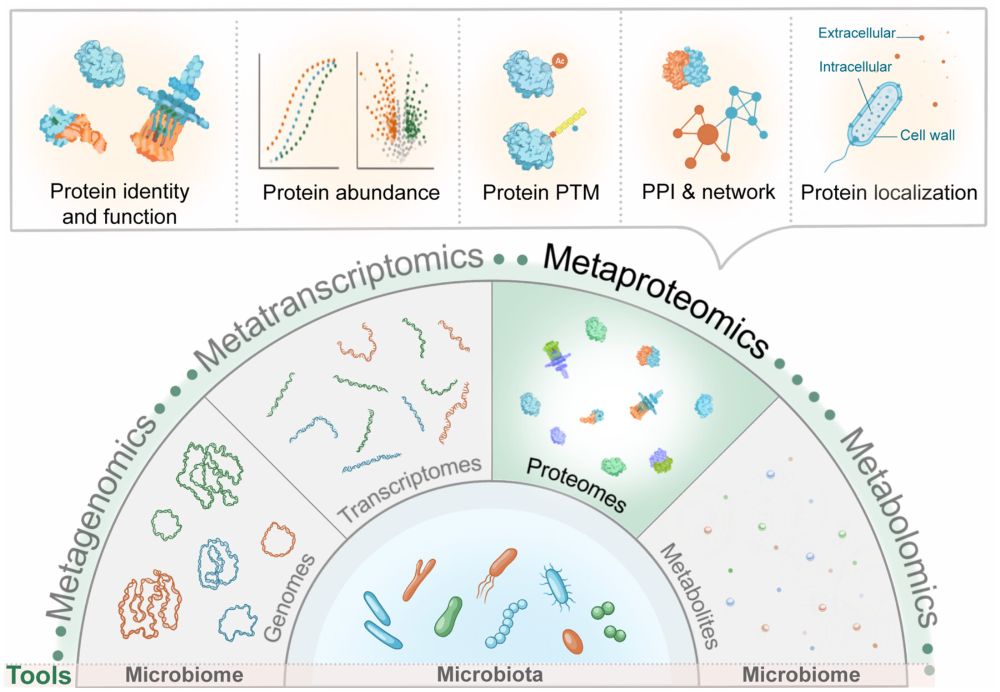
onlinelibrary.wiley.com/doi/full/10.... #Bioinformatics #microbiome #metaproteomics #proteomics #MS
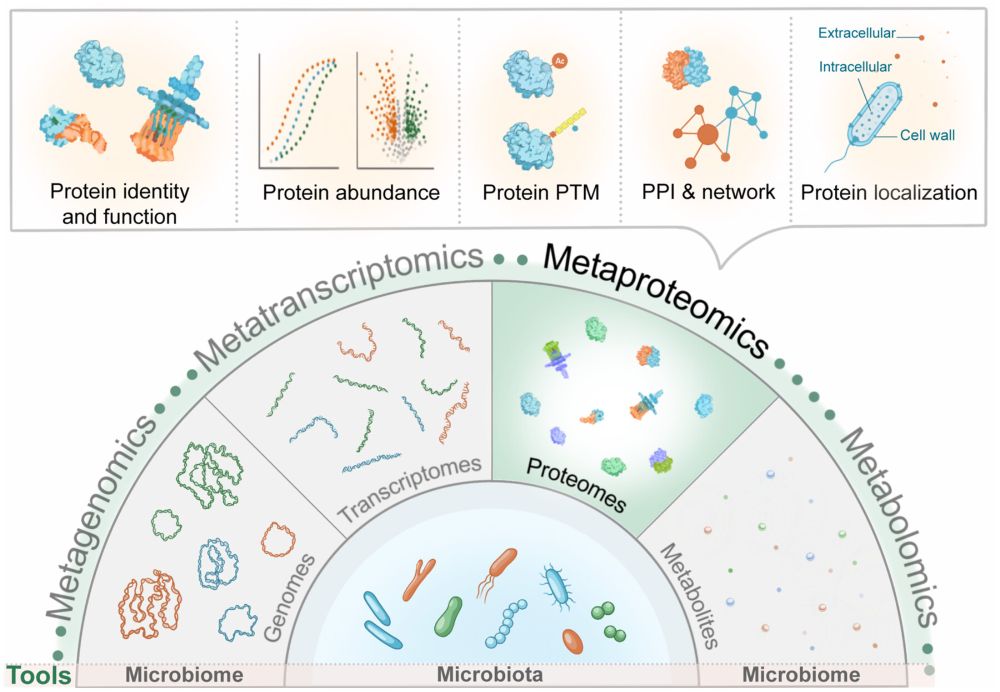
#microbiome #PrecisionNutrition #metaproteomics #metabolics #metagenomics

#microbiome #PrecisionNutrition #metaproteomics #metabolics #metagenomics
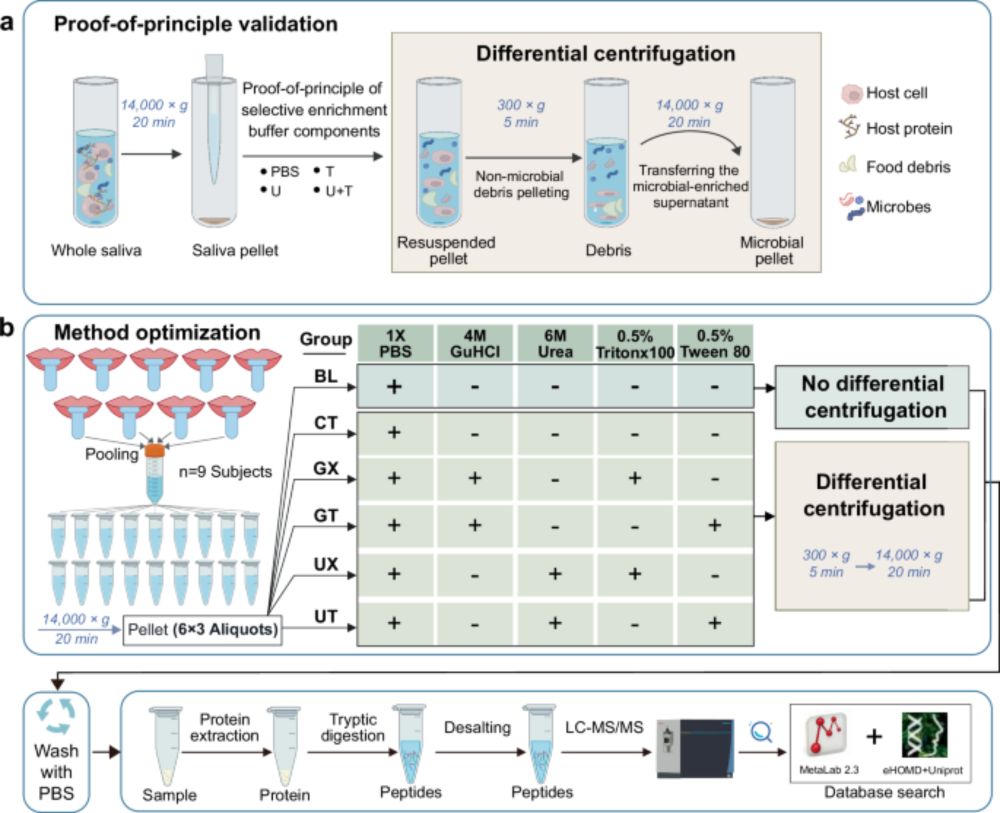

microbiolly.wordpress.com/2025/03/19/p...
microbiolly.wordpress.com/2025/03/19/p...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
🌱 We developed PhyloFunc, a beta diversity metric that integrates phylogeny into functional distance calculations, offering a more ecologically meaningful way to compare microbiomes.
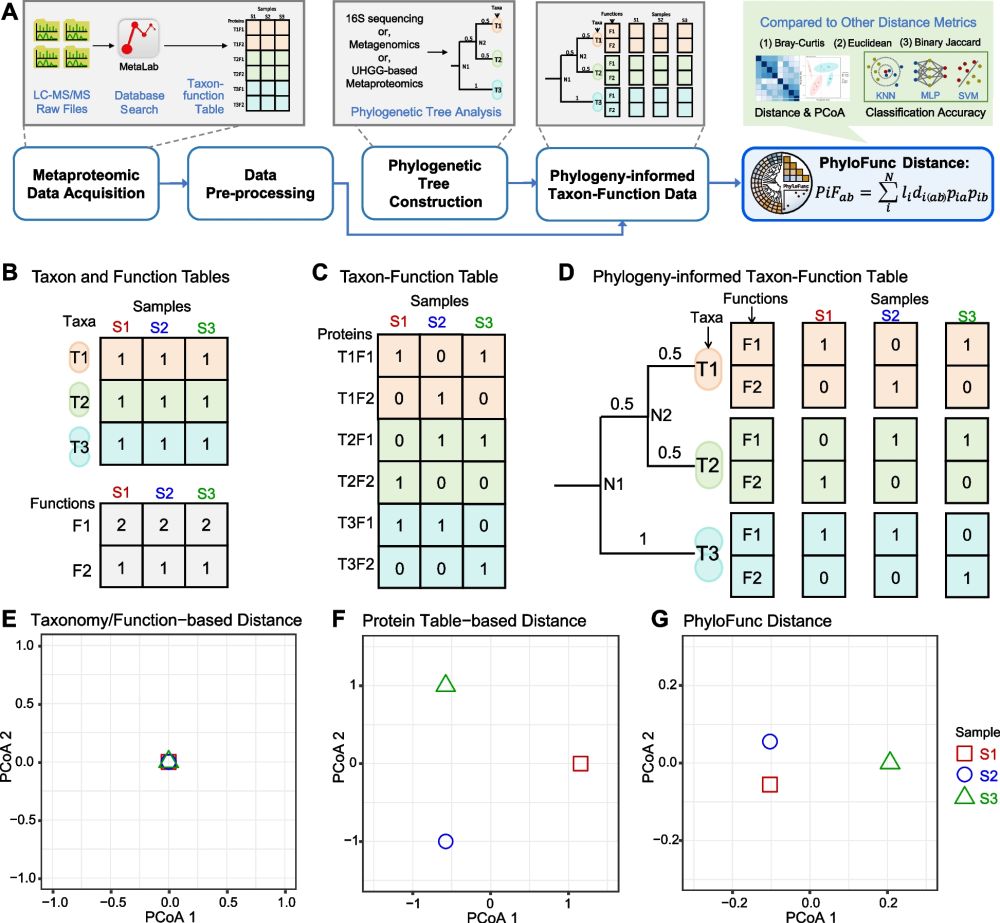
🌱 We developed PhyloFunc, a beta diversity metric that integrates phylogeny into functional distance calculations, offering a more ecologically meaningful way to compare microbiomes.

chemrxiv.org/engage/chemr...

chemrxiv.org/engage/chemr...

