
https://ninjani.github.io/
Pre-print: www.biorxiv.org/content/10.1...
🧵👇(1/n)

mirdita.org

mirdita.org
Thread below - but first some links:
A longer take:
fraserlab.com/2025/12/29/k...
Preprint:
www.biorxiv.org/content/10.6...
Thread below - but first some links:
A longer take:
fraserlab.com/2025/12/29/k...
Preprint:
www.biorxiv.org/content/10.6...
We present OpenStructure's powerful scoring capabilities, used to assess predictionsin CAMEO and CASP.
Read the full study here:
🔗 doi.org/10.1038/s415...
#StructuralBiology #Bioinformatics #OpenStructure #CASP #CAMEO #ProteinStructure
We present OpenStructure's powerful scoring capabilities, used to assess predictionsin CAMEO and CASP.
Read the full study here:
🔗 doi.org/10.1038/s415...
#StructuralBiology #Bioinformatics #OpenStructure #CASP #CAMEO #ProteinStructure
Pre-print: www.biorxiv.org/content/10.1...
🧵👇(1/n)

Day 2/31
Prompt WEAVE
N-terminal domain of a Fibrion - a building block of silk fiber produced by silkworms.
Pdb: 3UA0
Next prompt is CROWN and I would love your suggestions!

Day 2/31
Prompt WEAVE
N-terminal domain of a Fibrion - a building block of silk fiber produced by silkworms.
Pdb: 3UA0
Next prompt is CROWN and I would love your suggestions!
~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...

~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...
“From bytes to binders: design, score and optimize” #bc2basel #posterprize
“From bytes to binders: design, score and optimize” #bc2basel #posterprize
@sib.swiss @biozentrum.unibas.ch
⬇️⬇️⬇️
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

@sib.swiss @biozentrum.unibas.ch
⬇️⬇️⬇️
www.nature.com/articles/s41...
www.nature.com/articles/s41...
📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.

📅 Sept 8, 2025 | Basel
💡 Talks + breakout sessions on #CASP #CAPRI #CAMEO & benchmarking for drug discovery
🎟️ Free registration (limited spots): lu.ma/ws9nu1xf
Join us to explore how benchmarking can drive breakthroughs in structure prediction.
Find out about molecular modelling to systems-level analyses and evolution, and more.
Submit your abstract by 26 Aug ➡️ s.embl.org/csb25-01-bl

Find out about molecular modelling to systems-level analyses and evolution, and more.
Submit your abstract by 26 Aug ➡️ s.embl.org/csb25-01-bl
We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6

AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
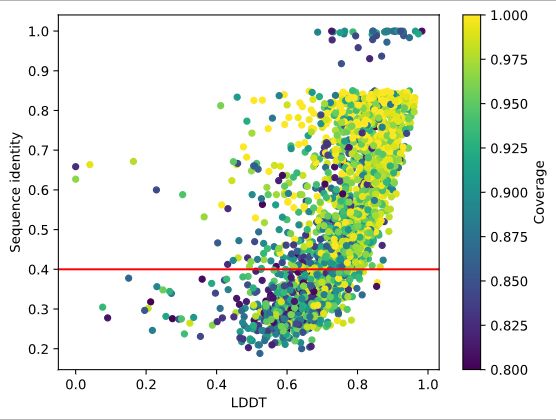
The summer call at @biozentrum.unibas.ch @unibas.ch is open until October 12, 2025.
www.biozentrum.unibas.ch/phd/internat...

The summer call at @biozentrum.unibas.ch @unibas.ch is open until October 12, 2025.
www.biozentrum.unibas.ch/phd/internat...
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

patwalters.github.io/Three-Papers...

patwalters.github.io/Three-Papers...

@torstenschwede.bsky.social @xavier.robin.info
Join us and register your server now!
cameo3d.org
@torstenschwede.bsky.social @xavier.robin.info


