
Evolutionary relationships and classification of protein domains.
https://cathdb.info
https://ted.cathdb.info




Congratulations to Jude Wells for Design in voxel space, decode in smiles space: Plixer generates drug-like molecules for protein pockets

We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.

A collaboration between the groups of @martinsteinegger.bsky.social , David Jones and Christine Orengo, we clustered AlphaFold Database and ESMatlas, a whopping 821 million proteins!
We reveal biome-specific groups & over 11k novel domain combinations.
The TED integration is set to enhance the interpretability and usability of #AlphaFold predictions. Is this useful in your work?
The TED integration is set to enhance the interpretability and usability of #AlphaFold predictions. Is this useful in your work?
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social
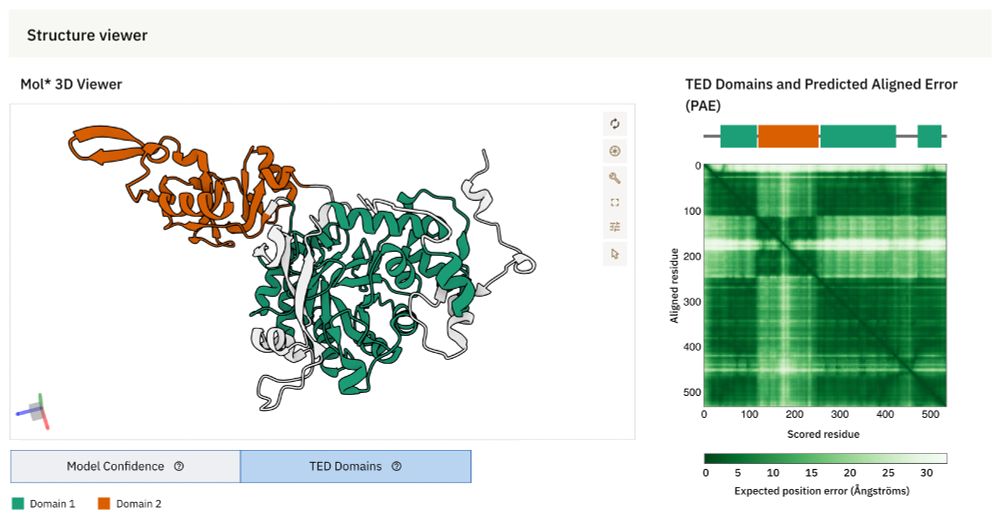
AlphaFold DB now integrates The Encyclopedia of Domains (TED) – a resource designed to systematically identify & classify structural domains within AlphaFold-predicted protein structures.
www.ebi.ac.uk/about/news/u...
@pdbeurope.bsky.social


Here’s a link to the manuscript in NAR.

Here’s a link to the manuscript in NAR.
Out now in Science, we are finally ready to make our classification of all domains in AFDB v4 available to all!
A joint effort with the Jones group at UCL Computer Science, here's a link to the article
www.science.org/doi/10.1126/...
1/6 🧵⤵️

Out now in Science, we are finally ready to make our classification of all domains in AFDB v4 available to all!
A joint effort with the Jones group at UCL Computer Science, here's a link to the article
www.science.org/doi/10.1126/...
1/6 🧵⤵️
We are CATH! We classify protein domains in homologous superfamilies from the Protein Data Bank and AlphaFold Database!
Follow us for our latest research!
We are CATH! We classify protein domains in homologous superfamilies from the Protein Data Bank and AlphaFold Database!
Follow us for our latest research!

