
Evolutionary relationships and classification of protein domains.
https://cathdb.info
https://ted.cathdb.info











6/6

6/6
zenodo.org/records/1390...
Here you can find per-chain and per-domain annotations, domain interaction data and model weights
4/6 🧵⤵️

zenodo.org/records/1390...
Here you can find per-chain and per-domain annotations, domain interaction data and model weights
4/6 🧵⤵️
3/6 🧵⤵️

3/6 🧵⤵️
Each of the 214M chains in TED is now being treated equally, identifying 365 million protein domains.
2/6 🧵⤵️
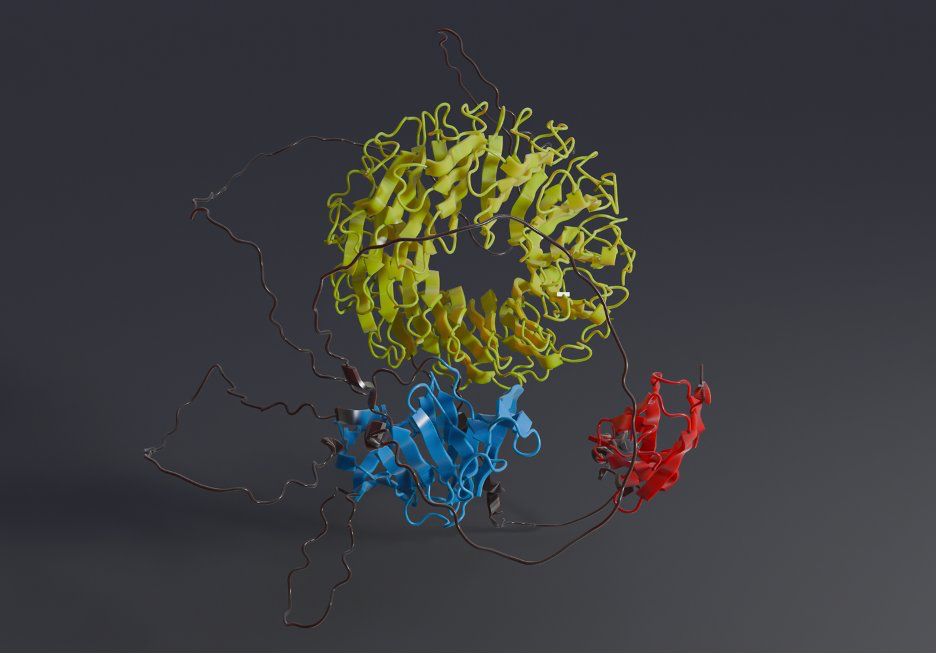
Each of the 214M chains in TED is now being treated equally, identifying 365 million protein domains.
2/6 🧵⤵️

