CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
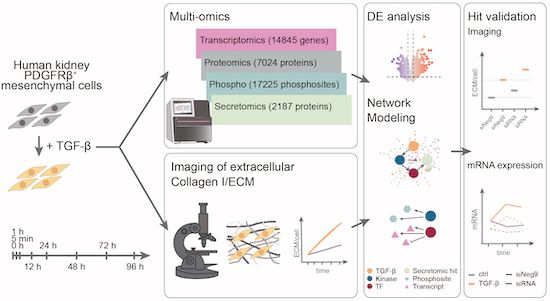

#Glycoproteins #Glycoforms #Glycoprofiling #DQGlyco #Glycotime
authors.elsevier.com/a/1l3k53S6Gf...
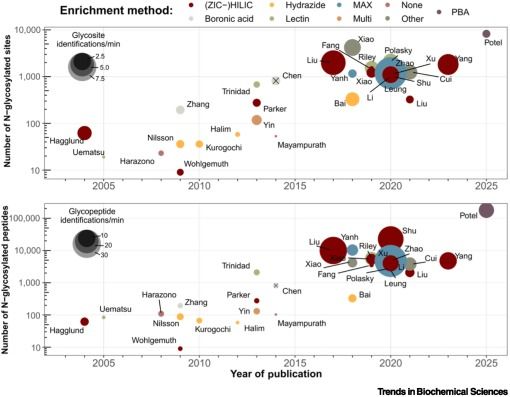
#Glycoproteins #Glycoforms #Glycoprofiling #DQGlyco #Glycotime
authors.elsevier.com/a/1l3k53S6Gf...

Huge kudos to all coauthors, especially the amazing glycoteam: Clement, @miraburtscher.bsky.social, Isabelle, and Misha (@savitski-lab.bsky.social)!
www.nature.com/articles/s41...
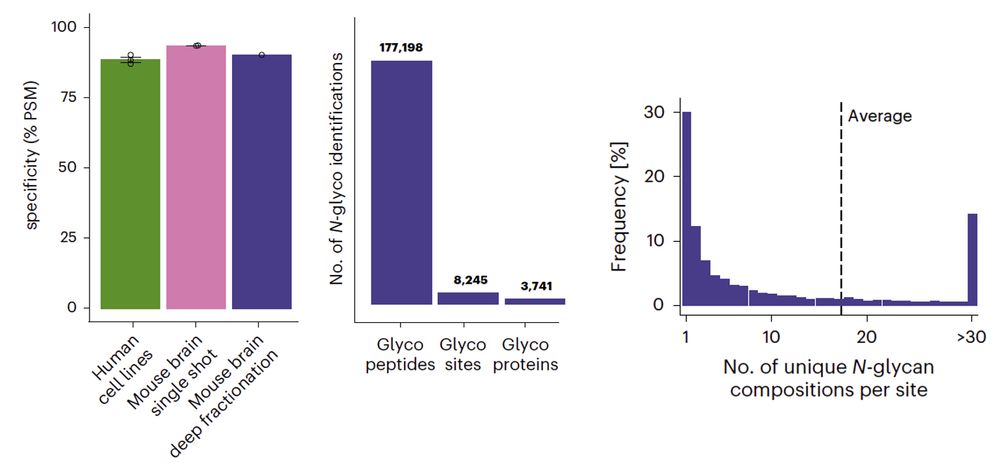
Huge kudos to all coauthors, especially the amazing glycoteam: Clement, @miraburtscher.bsky.social, Isabelle, and Misha (@savitski-lab.bsky.social)!
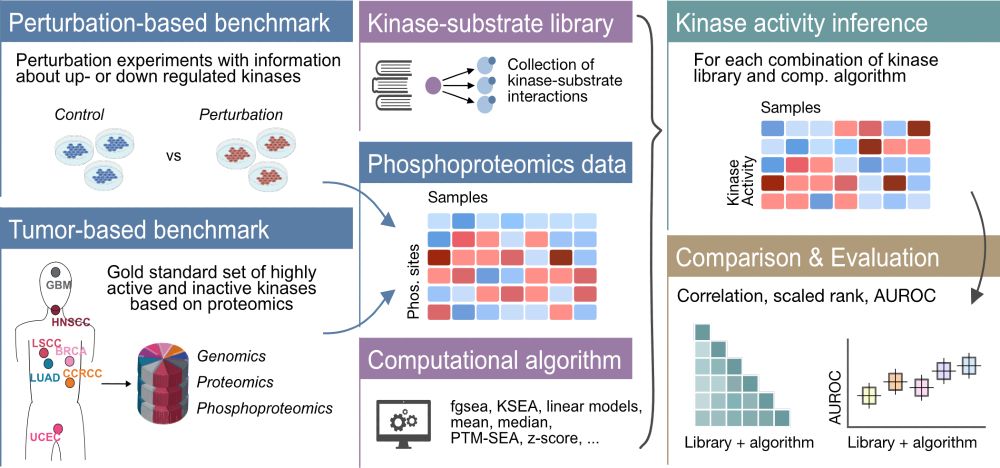
In their study, the scientists describe a new method to study glycosylation systematically & quantitatively, leading to new biological insights. 🧪🧠📈
www.embl.org/news/science...
In their study, the scientists describe a new method to study glycosylation systematically & quantitatively, leading to new biological insights. 🧪🧠📈
www.embl.org/news/science...
www.nature.com/articles/s41...


