
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape

We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape


www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵


www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇

🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
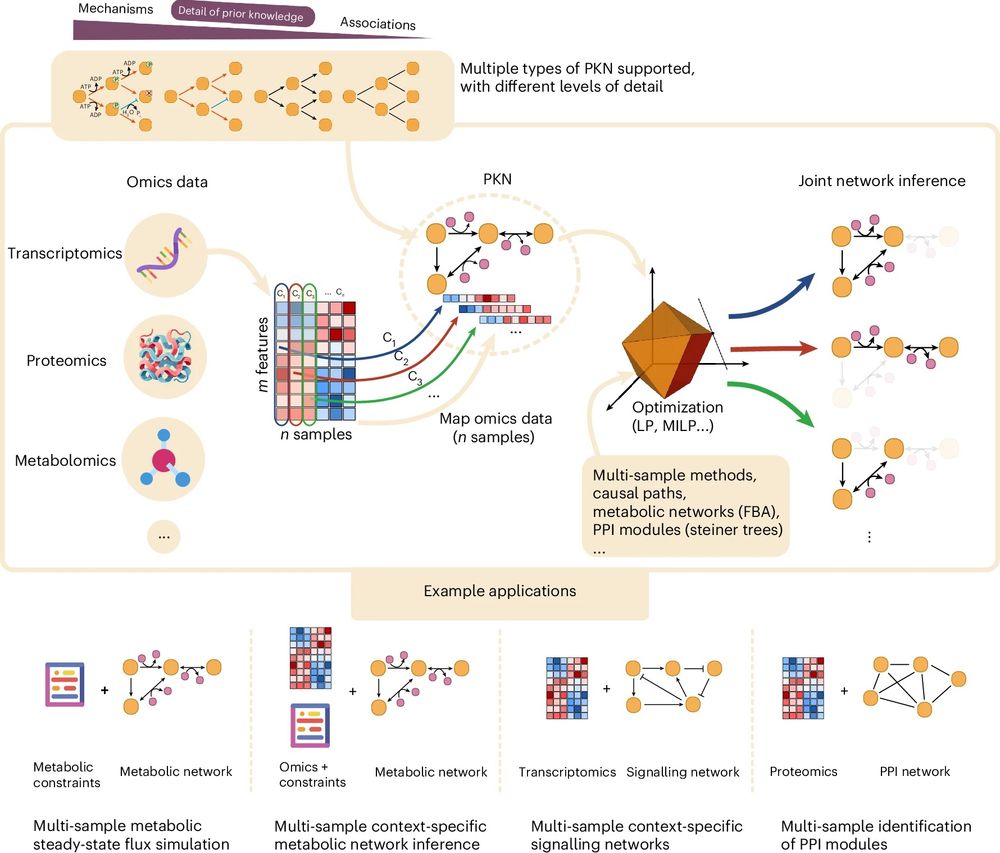
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪

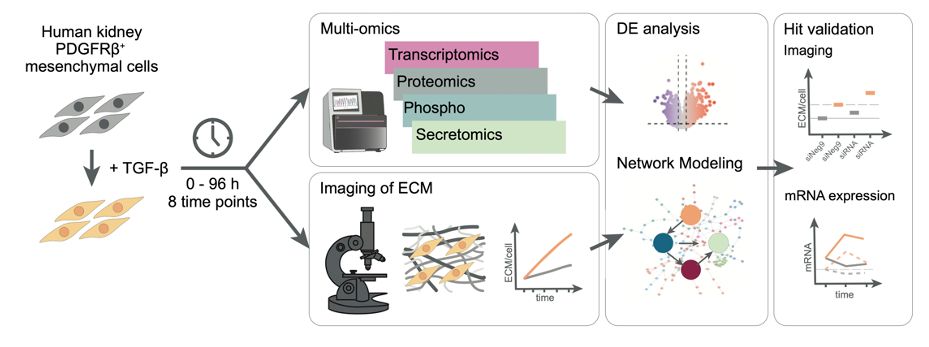

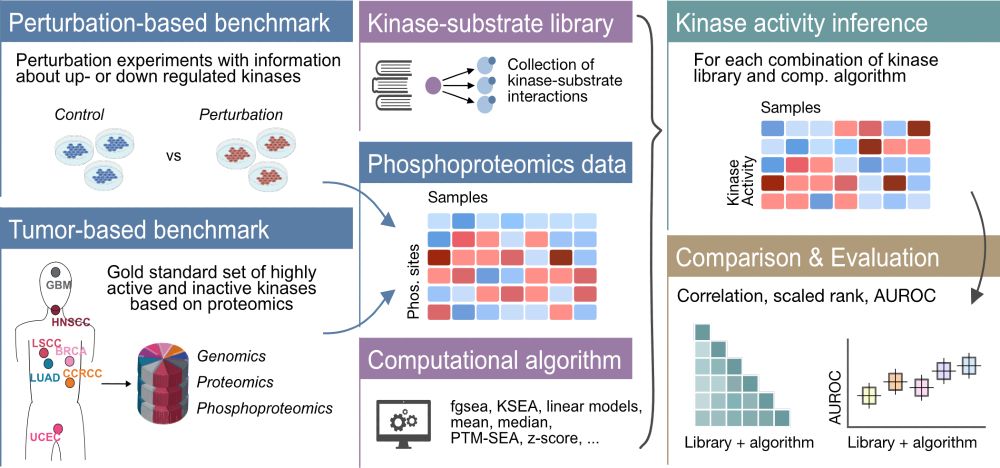

www.cell.com/cell-systems...
Big thanks to @juliosaezrod.bsky.social and @cp-cellsystems.bsky.social for the opportunity to contribute!

www.cell.com/cell-systems...
Big thanks to @juliosaezrod.bsky.social and @cp-cellsystems.bsky.social for the opportunity to contribute!

www.nature.com/articles/s41...

www.nature.com/articles/s41...
#Glycoproteins #Glycoforms #Glycoprofiling #DQGlyco #Glycotime
authors.elsevier.com/a/1l3k53S6Gf...
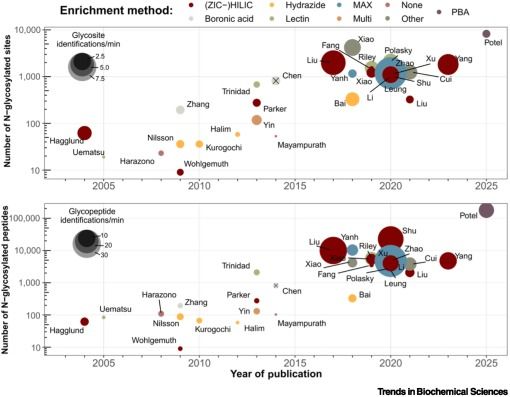
#Glycoproteins #Glycoforms #Glycoprofiling #DQGlyco #Glycotime
authors.elsevier.com/a/1l3k53S6Gf...

We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
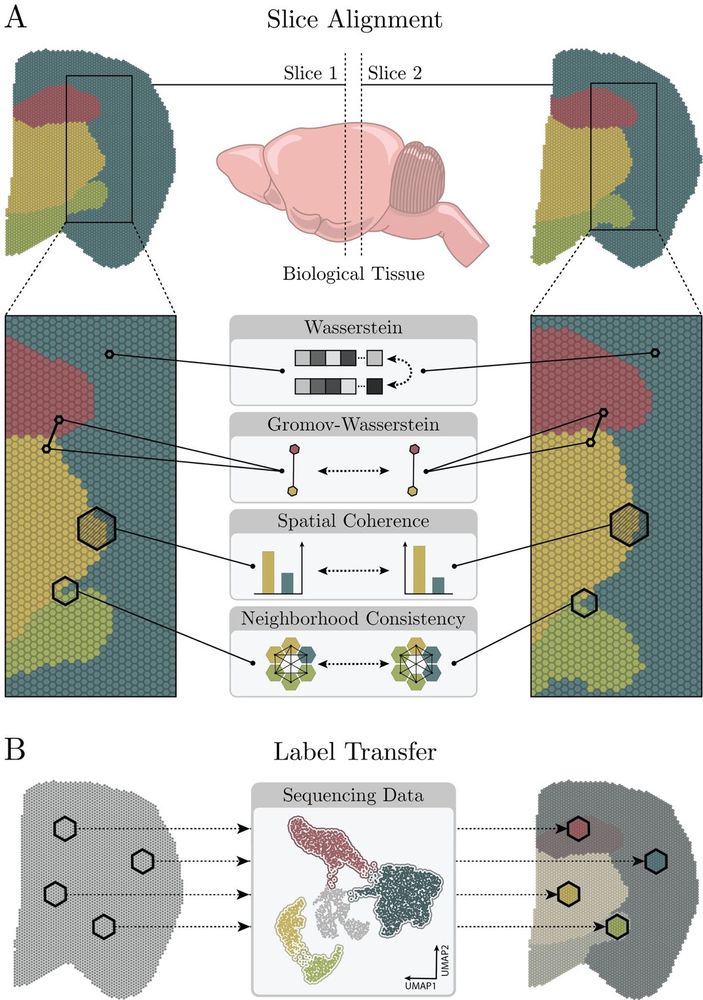
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇

We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇



