Julio Saez-Rodriguez
@juliosaezrod.bsky.social
Computational Biologist, Head of research @ebi.embl.org, part-time
Heidelberg University, codirector DREAM challenges. For group's activities, see @saezlab.bsky.social
Heidelberg University, codirector DREAM challenges. For group's activities, see @saezlab.bsky.social
Reposted by Julio Saez-Rodriguez
Our revised consensus transcriptional patient map of human heart failure across patient cohorts and single-cell and bulk technologies is now published @natcomms.nature.com www.nature.com/articles/s41...
What are the key disrupted multicellular processes in heart failure? In our new work we combine 23 years of molecular data with recent single-cell atlases to draw a cross-study patient map
doi.org/10.1101/2024...
doi.org/10.1101/2024...

November 6, 2025 at 12:10 PM
Our revised consensus transcriptional patient map of human heart failure across patient cohorts and single-cell and bulk technologies is now published @natcomms.nature.com www.nature.com/articles/s41...
Reposted by Julio Saez-Rodriguez
‼️ Deadline extension alert ‼️
The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!
The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!

October 17, 2025 at 11:37 AM
‼️ Deadline extension alert ‼️
The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!
The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!
Reposted by Julio Saez-Rodriguez
Very happy to see this work finally online 🥳
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
September 30, 2025 at 12:23 PM
Very happy to see this work finally online 🥳
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
Reposted by Julio Saez-Rodriguez
I'm presenting #CIRCE, a Python package to infer co-accessible DNA region networks 🧬
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️

September 30, 2025 at 12:23 PM
I'm presenting #CIRCE, a Python package to infer co-accessible DNA region networks 🧬
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Reposted by Julio Saez-Rodriguez
We are hiring a Postdoctoral Fellow in Computational Biology at EMBL-EBI (Cambridge, UK). Focus: methods to study cell–cell communication from sc/spatial omics data (building on LIANA+ and NicheNet), in collab with @yvansaeys.bsky.social VIB/Ghent.
Details & apply by 13/10/25: tinyurl.com/4shdw8dk
Details & apply by 13/10/25: tinyurl.com/4shdw8dk

Current Vacancies
Whether you're a scientist, IT specialist, accountant or administrator, you'll help us tackle the challenges of improving human health & biodiversity in the face of climate change on a global scal...
tinyurl.com
September 26, 2025 at 10:16 AM
We are hiring a Postdoctoral Fellow in Computational Biology at EMBL-EBI (Cambridge, UK). Focus: methods to study cell–cell communication from sc/spatial omics data (building on LIANA+ and NicheNet), in collab with @yvansaeys.bsky.social VIB/Ghent.
Details & apply by 13/10/25: tinyurl.com/4shdw8dk
Details & apply by 13/10/25: tinyurl.com/4shdw8dk
Reposted by Julio Saez-Rodriguez
🥁 Check out our new preprint on OmniPath, the prior knowledge resource for #SystemsBiology, and its brand-new OmniPath Explorer web app! 🥳
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️

September 17, 2025 at 1:10 PM
🥁 Check out our new preprint on OmniPath, the prior knowledge resource for #SystemsBiology, and its brand-new OmniPath Explorer web app! 🥳
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
Reposted by Julio Saez-Rodriguez
🚨 New preprint
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape

September 19, 2025 at 7:45 AM
🚨 New preprint
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
Reposted by Julio Saez-Rodriguez
Introducing ParTIpy, a python package for Pareto Task Inference that scales to large-scale datasets, including single-cell and spatial transcriptomics.
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

September 15, 2025 at 8:40 AM
Introducing ParTIpy, a python package for Pareto Task Inference that scales to large-scale datasets, including single-cell and spatial transcriptomics.
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
Reposted by Julio Saez-Rodriguez
Meet our newest Research Group Leader: Jess Ewald @ewaldlab.org 🇨🇦
Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Welcome: Jess Ewald
Jess Ewald, Research Group Leader at EMBL-EBI, uses cell profiling and AI to find and characterise chemical hazards to humans and ecosysystems.
www.ebi.ac.uk
September 3, 2025 at 9:07 AM
Meet our newest Research Group Leader: Jess Ewald @ewaldlab.org 🇨🇦
Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Reposted by Julio Saez-Rodriguez
Welcome @timcoorens.bsky.social 🇳🇱, our new Research Group Leader.
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Welcome: Tim Coorens
EMBL-EBI’s newest Research Group Leader is investigating how somatic mutations reveal the hidden histories of human cells.
www.ebi.ac.uk
August 12, 2025 at 9:04 AM
Welcome @timcoorens.bsky.social 🇳🇱, our new Research Group Leader.
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬
@embl.org PhD programme just open - many interesting projects, including two with us @saezlab.bsky.social @ebi.embl.org - one on #host-microbiome interactions using #spatial omics, and one on #AI in #proteomics 👇
📢 Recruitment for the EMBL International PhD Programme is officially open!
At EMBL, we train young scientists to become skilled and creative future leaders in academia and other sectors. Join us and get a head start on your career in life sciences!
bit.ly/47mmiFF
At EMBL, we train young scientists to become skilled and creative future leaders in academia and other sectors. Join us and get a head start on your career in life sciences!
bit.ly/47mmiFF

September 2, 2025 at 6:55 PM
@embl.org PhD programme just open - many interesting projects, including two with us @saezlab.bsky.social @ebi.embl.org - one on #host-microbiome interactions using #spatial omics, and one on #AI in #proteomics 👇
Reposted by Julio Saez-Rodriguez
👋 Time to say goodbye to another one of our PhD students, @smuellerdott.bsky.social. She was working on the integration of phosphoproteomics and transcriptomics data and will now join the Computational Oncology Team at Merck. We wish her all the best for this new chapter! 🚀 ✨


September 1, 2025 at 5:35 AM
👋 Time to say goodbye to another one of our PhD students, @smuellerdott.bsky.social. She was working on the integration of phosphoproteomics and transcriptomics data and will now join the Computational Oncology Team at Merck. We wish her all the best for this new chapter! 🚀 ✨
Reposted by Julio Saez-Rodriguez
We present our MetaProViz #Rpackage for #metabolomics analysis & prior knowledge integration to generate mechanistic hypotheses on how metabolic changes affect metabolite classes, pathways & environment interaction
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️

August 25, 2025 at 7:07 AM
We present our MetaProViz #Rpackage for #metabolomics analysis & prior knowledge integration to generate mechanistic hypotheses on how metabolic changes affect metabolite classes, pathways & environment interaction
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
Reposted by Julio Saez-Rodriguez
Glad to contribute to this collaborative community platform.
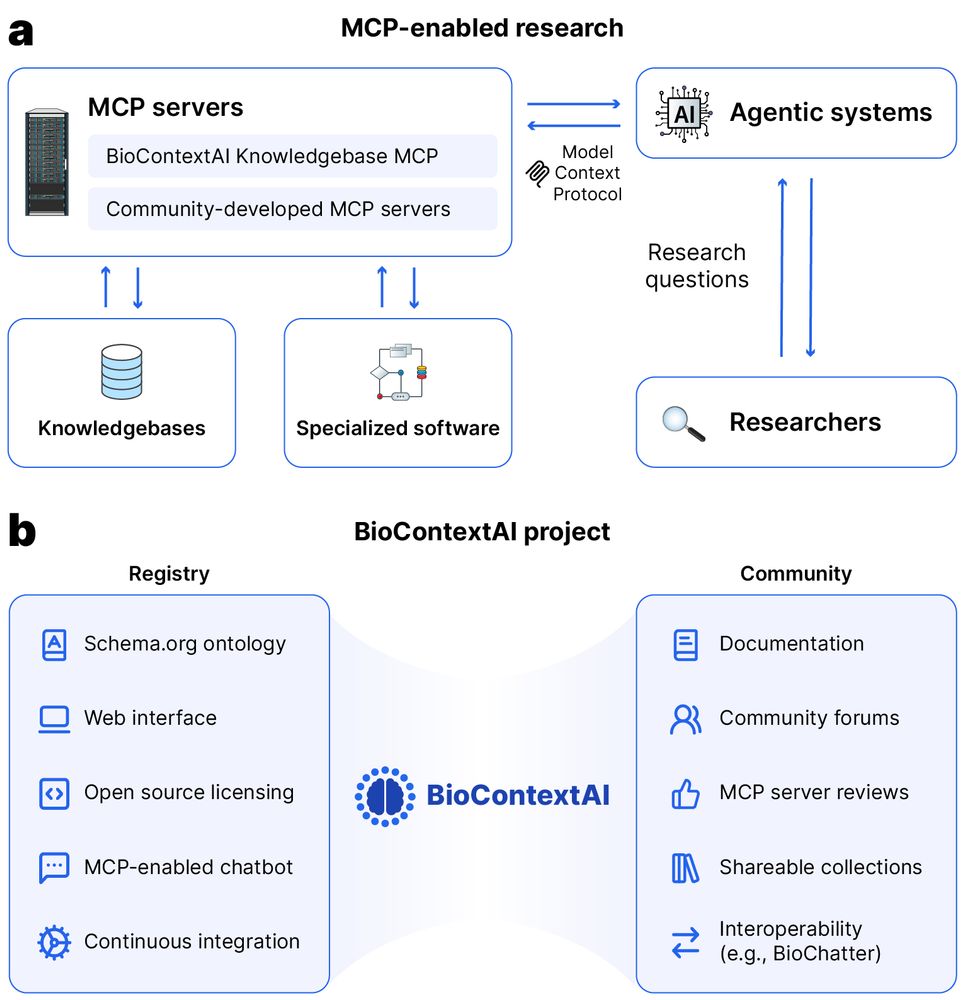
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
July 31, 2025 at 7:37 AM
Glad to contribute to this collaborative community platform.
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
Reposted by Julio Saez-Rodriguez
👀 we are involved in projects 1 & 6 on the list:
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
Recruitment is now open for the EMBL-EBI–Sanger Postdoctoral Programme.
ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob
ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob

August 14, 2025 at 9:05 AM
👀 we are involved in projects 1 & 6 on the list:
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
Reposted by Julio Saez-Rodriguez
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...

July 29, 2025 at 12:41 PM
Preprint alert 🚨 Do you use chatbots in your work or even build MCP servers and agentic systems yourself?
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
Reposted by Julio Saez-Rodriguez
Our group @ebi.embl.org is a host for the ARISE2 MSCA-funded post-doc fellowship, in collab with @bioimagearchive.bsky.social and @scilifelab.se (esp. A Mezger and S. Giacomello) to support the development of next-gen spatial data infrastructures. Apply by 30/09
www.embl.org/training/ari...
www.embl.org/training/ari...
ARISE2
Career Accelerator for Research Infrastructure Scientists
www.embl.org
July 14, 2025 at 9:59 AM
Our group @ebi.embl.org is a host for the ARISE2 MSCA-funded post-doc fellowship, in collab with @bioimagearchive.bsky.social and @scilifelab.se (esp. A Mezger and S. Giacomello) to support the development of next-gen spatial data infrastructures. Apply by 30/09
www.embl.org/training/ari...
www.embl.org/training/ari...
Reposted by Julio Saez-Rodriguez
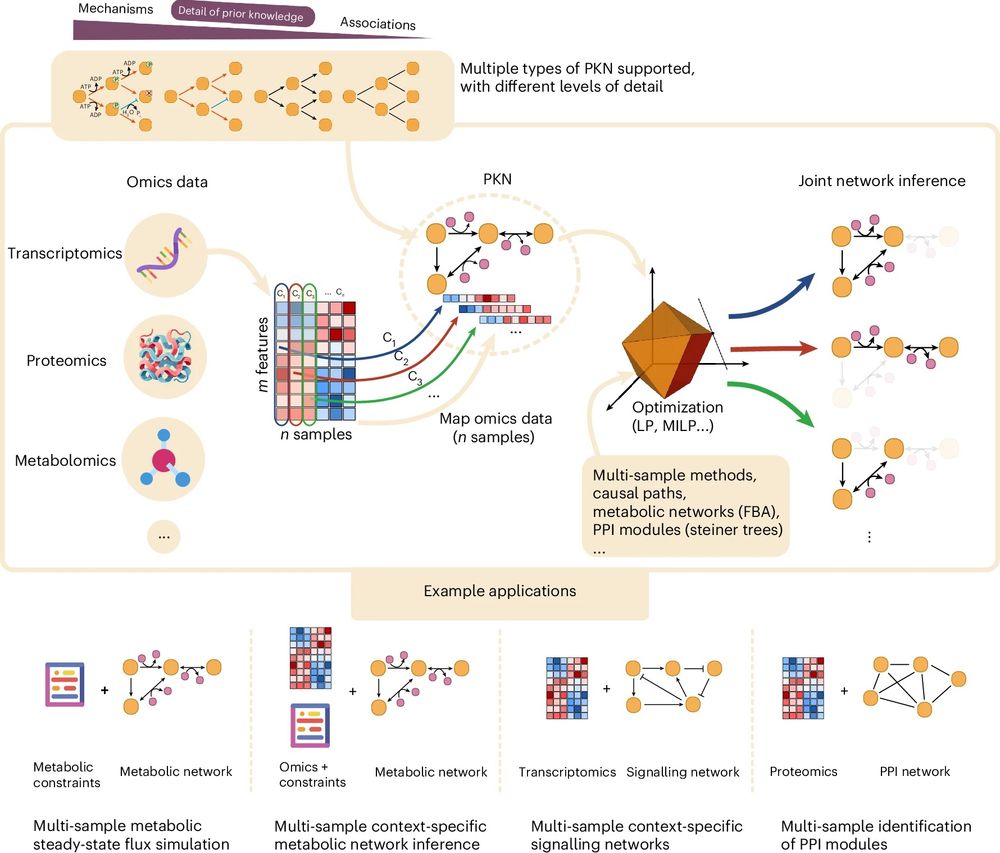
🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is published in peer reviewed form
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
July 22, 2025 at 3:23 PM
🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is published in peer reviewed form
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
Reposted by Julio Saez-Rodriguez
join us at ISMB iscb.bsky.social & @dreamchallenges.bsky.social - benchmarking foundation models w. @anshulkundaje.bsky.social @oldbap.bsky.social G. Stolovitzky @bowang87.bsky.social @mariabrbic.bsky.social K. Kalantar, J. Guinney supported by @ebi.embl.org @cziscience.bsky.social Tempus AI
May 16, 2025 at 11:23 AM
join us at ISMB iscb.bsky.social & @dreamchallenges.bsky.social - benchmarking foundation models w. @anshulkundaje.bsky.social @oldbap.bsky.social G. Stolovitzky @bowang87.bsky.social @mariabrbic.bsky.social K. Kalantar, J. Guinney supported by @ebi.embl.org @cziscience.bsky.social Tempus AI
Reposted by Julio Saez-Rodriguez
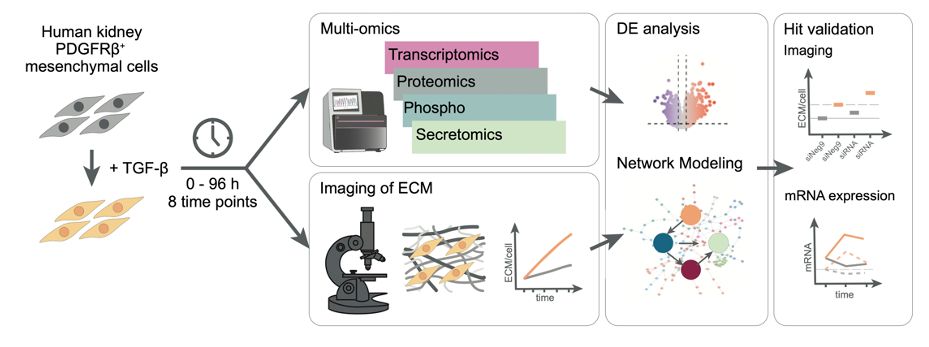
The final version of our multi-omics study on kidney fibrosis is out now (tinyurl.com/kidneyfibMSB). Together w/ Pepperkok + Savitski labs @embl.org, we present a time-resolved #multiomics + computational network modeling approach in combination w/ phenotypic assays to study #kidneyfibrosis
June 27, 2025 at 1:29 PM
The final version of our multi-omics study on kidney fibrosis is out now (tinyurl.com/kidneyfibMSB). Together w/ Pepperkok + Savitski labs @embl.org, we present a time-resolved #multiomics + computational network modeling approach in combination w/ phenotypic assays to study #kidneyfibrosis
Reposted by Julio Saez-Rodriguez
👋 Wishing @paubadiam.bsky.social all the best as he leaves our lab to join the Stanford Department of Genetics in @anshulkundaje.bsky.social lab! Looking forward to his future contributions to understanding gene regulation through sequence-to-function deep learning models 💻

June 23, 2025 at 11:26 AM
👋 Wishing @paubadiam.bsky.social all the best as he leaves our lab to join the Stanford Department of Genetics in @anshulkundaje.bsky.social lab! Looking forward to his future contributions to understanding gene regulation through sequence-to-function deep learning models 💻
I wrote a short piece on how becoming an IgA Nephropathy patient has changed my perspective on biomedical research, developing an appreciation for the challenges in data interpretability & availability and the importance of patient engagement www.nature.com/articles/s41...

Leveraging data as a patient–scientist: frustrations and opportunities - Nature Reviews Nephrology
The transition from data scientist to patient–scientist has given me new perspectives into clinical research and strengthened my commitment to open science. Although limitations on data availability h...
www.nature.com
June 23, 2025 at 6:19 AM
I wrote a short piece on how becoming an IgA Nephropathy patient has changed my perspective on biomedical research, developing an appreciation for the challenges in data interpretability & availability and the importance of patient engagement www.nature.com/articles/s41...
Reposted by Julio Saez-Rodriguez
🔧 decoupler 2.0.6 is out thanks to @paubadiam.bsky.social, now as a core @scverse.bsky.social package scverse.org
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
June 12, 2025 at 6:36 AM
🔧 decoupler 2.0.6 is out thanks to @paubadiam.bsky.social, now as a core @scverse.bsky.social package scverse.org
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
Reposted by Julio Saez-Rodriguez
In this Voices article, our @martingarridorc.bsky.social & @juliosaezrod.bsky.social discuss key strategies for building context-specific molecular networks: knowledge and data-driven methods, + future directions in the field bridging to structures and perturbations tinyurl.com/z8t6yych

What is the current bottleneck in mapping molecular interaction networks?
Network biologists today have access to a rich assortment of interaction networks
produced by assays such as affinity purification-mass spectrometry (AP-MS), yeast
two-hybrid (Y2H) screening, co-fract...
www.cell.com
May 30, 2025 at 6:27 AM
In this Voices article, our @martingarridorc.bsky.social & @juliosaezrod.bsky.social discuss key strategies for building context-specific molecular networks: knowledge and data-driven methods, + future directions in the field bridging to structures and perturbations tinyurl.com/z8t6yych
Reposted by Julio Saez-Rodriguez
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab @bcmhouston.bsky.social, is now out @natcomms.nature.com 🔬 tinyurl.com/4twuc6z4
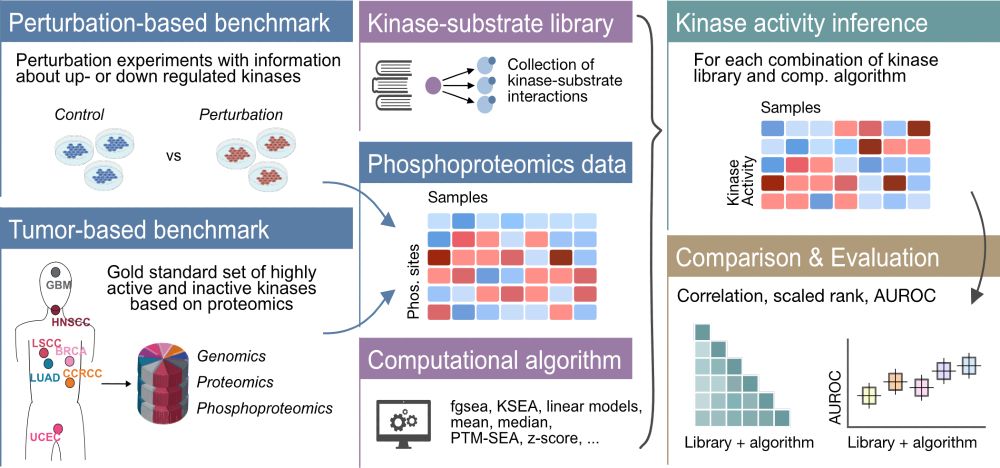
May 23, 2025 at 7:38 AM
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab @bcmhouston.bsky.social, is now out @natcomms.nature.com 🔬 tinyurl.com/4twuc6z4

