computational scientific discovery, biomedicine, spatial omics
Using spatial proteomics on ~500 tumors, we found distinct trajectories from early to late stage, involving the whole tumor microenvironment and its metabolic state.
📄 Preprint: arxiv.org/abs/2510.05083
#SpatialBiology #CRC #ImageBasedProfiling
1/5

Using spatial proteomics on ~500 tumors, we found distinct trajectories from early to late stage, involving the whole tumor microenvironment and its metabolic state.
📄 Preprint: arxiv.org/abs/2510.05083
#SpatialBiology #CRC #ImageBasedProfiling
1/5
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
🧵Walk-through thread below ⬇️
Well said @tanevski.bsky.social "Cancer is a spatial disease-spatialomics is the future of cancer science"!
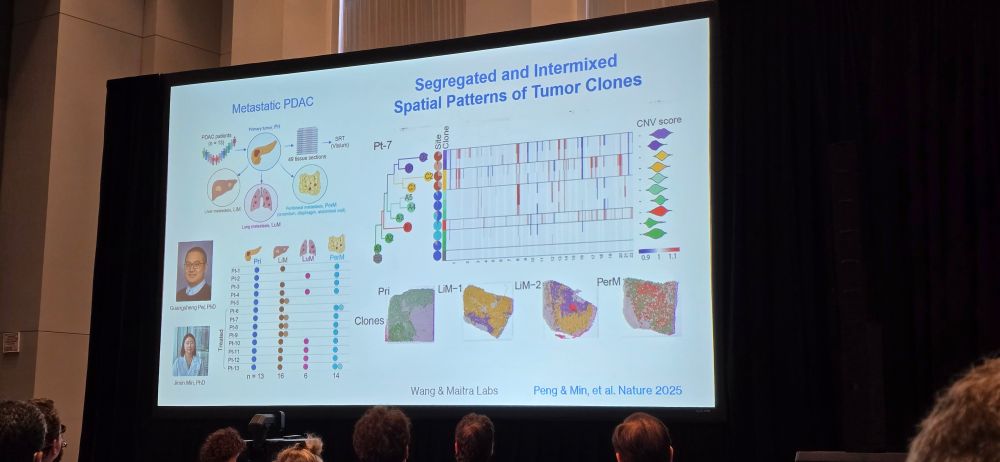
Wonderful composite spatial data from Linghua Wang @mdanderson.bsky.social
www.nature.com/articles/s41...

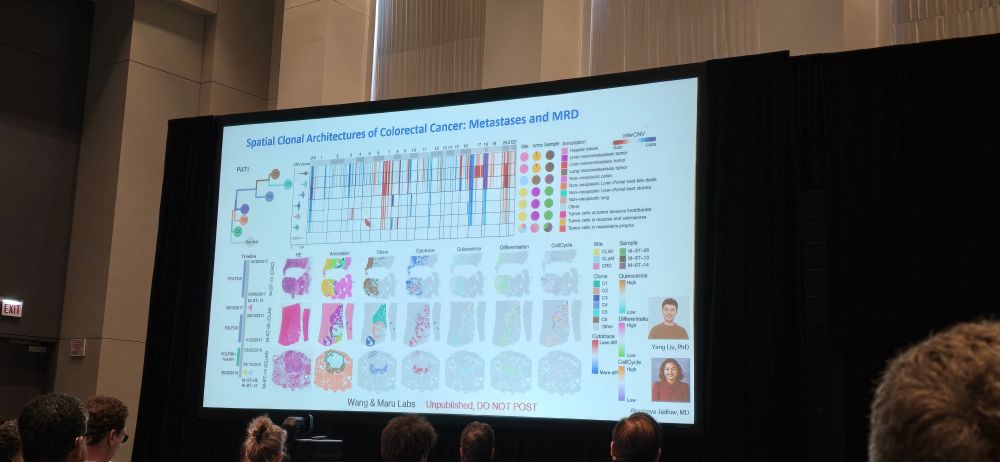
Well said @tanevski.bsky.social "Cancer is a spatial disease-spatialomics is the future of cancer science"!
Wonderful composite spatial data from Linghua Wang @mdanderson.bsky.social
www.nature.com/articles/s41...
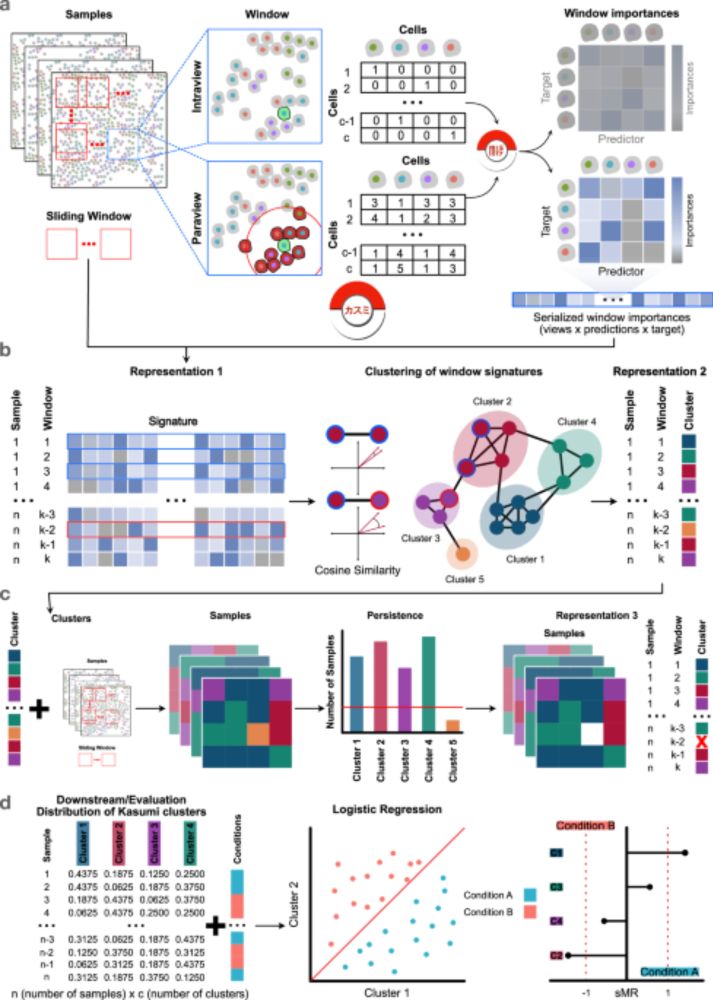
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
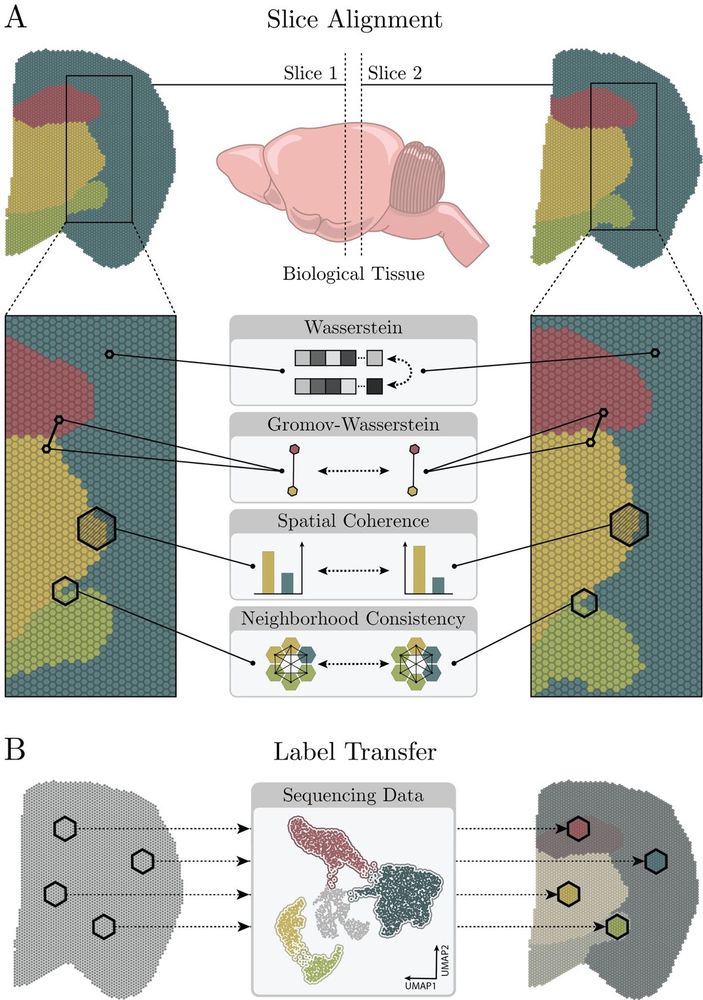
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
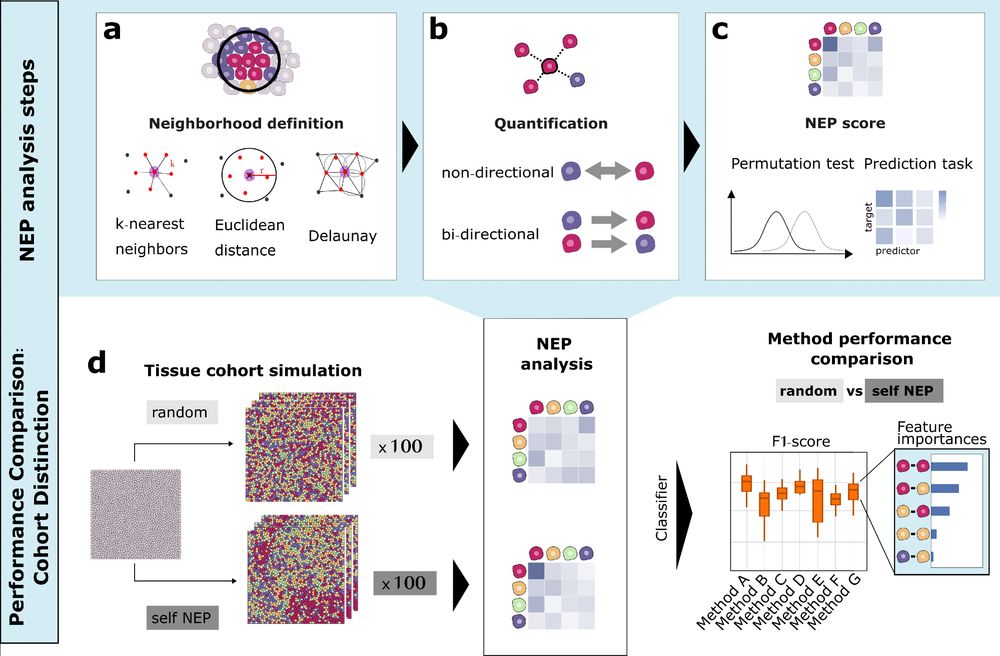
Ever wondered how to best quantify cell-cell neighbor preferences in tissues?
We compared 9+ neighbor preference (NEP) methods for analysing spatial omics data and propose a novel approach that combines the most relevant analysis features which we call COZI 🔬✨
Read more: doi.org/10.1101/2025...