Previously at Genentech, UBC and BITS Pilani
https://scholar.google.com/citations?user=4yUtALcAAAAJ&hl=en&oi=ao
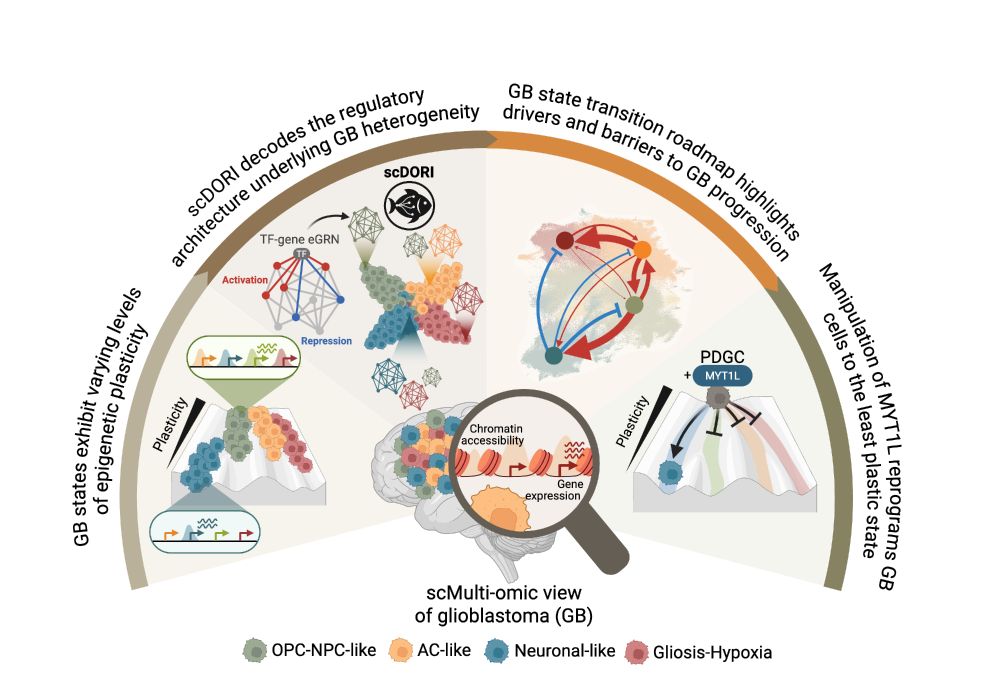
Delighted to have co-conceived this project with lead author @danaivagiaki.bsky.social
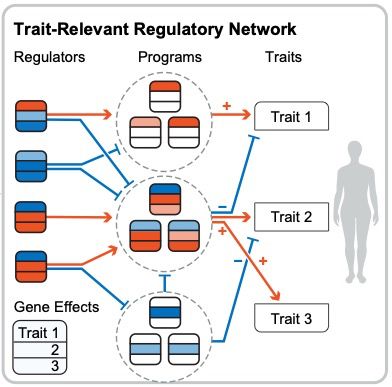
LIVI enables systematic discovery of genetic effects on gene regulatory networks in population-scale scRNA cohorts. Thread below 🧵👇🏼
Delighted to have co-conceived this project with lead author @danaivagiaki.bsky.social
LIVI enables systematic discovery of genetic effects on gene regulatory networks in population-scale scRNA cohorts. Thread below 🧵👇🏼

Will be presenting posters on my latest work on personalized gene expression prediction and gene regulatory network inference

Will be presenting posters on my latest work on personalized gene expression prediction and gene regulatory network inference
@gosiatrynka.bsky.social
@dgmacarthur.bsky.social
@bpasaniuc.bsky.social
@tuuliel.bsky.social
@hilarycmartin.bsky.social
@sashagusevposts.bsky.social
@zkutalik.bsky.social
@mashaals.bsky.social
@alemedinarivera.bsky.social

genomebiology.biomedcentral.com/articles/10....
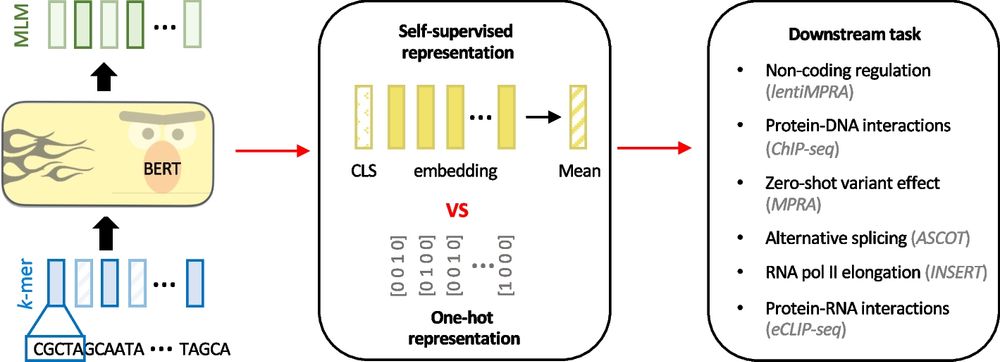
genomebiology.biomedcentral.com/articles/10....

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵


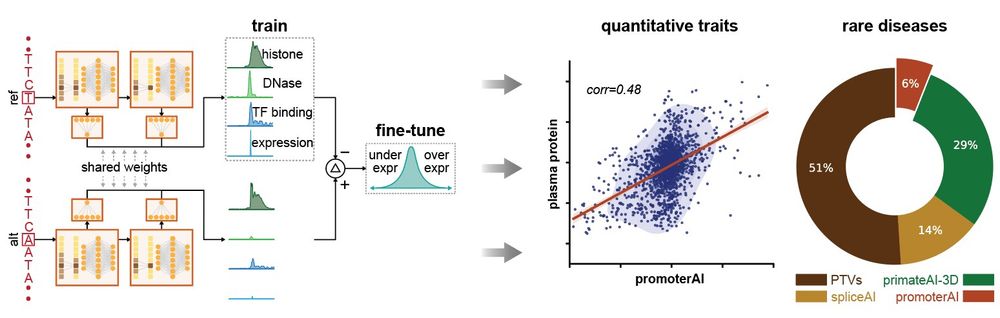
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
www.nature.com/articles/s41...
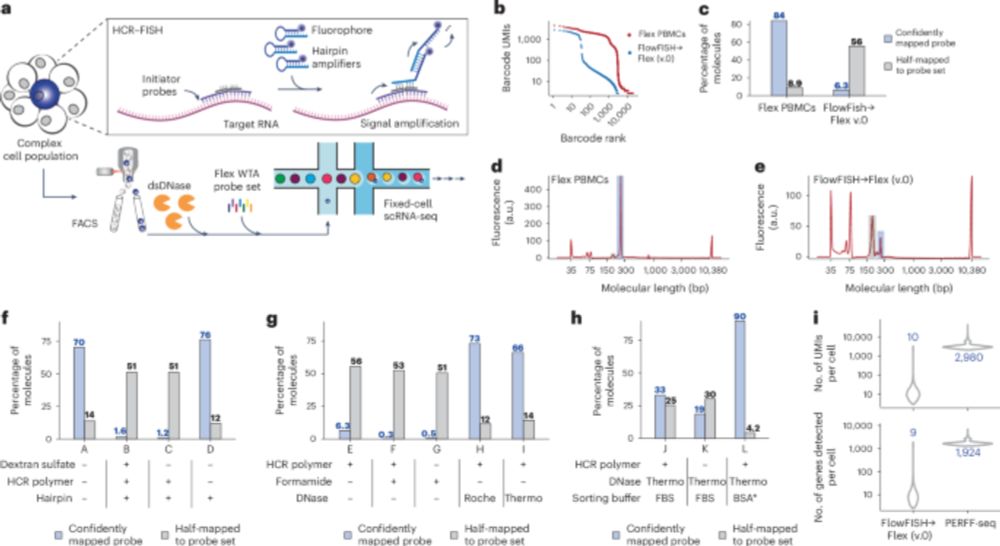
www.nature.com/articles/s41...
Can't wait to dive into the details
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
Can't wait to dive into the details
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
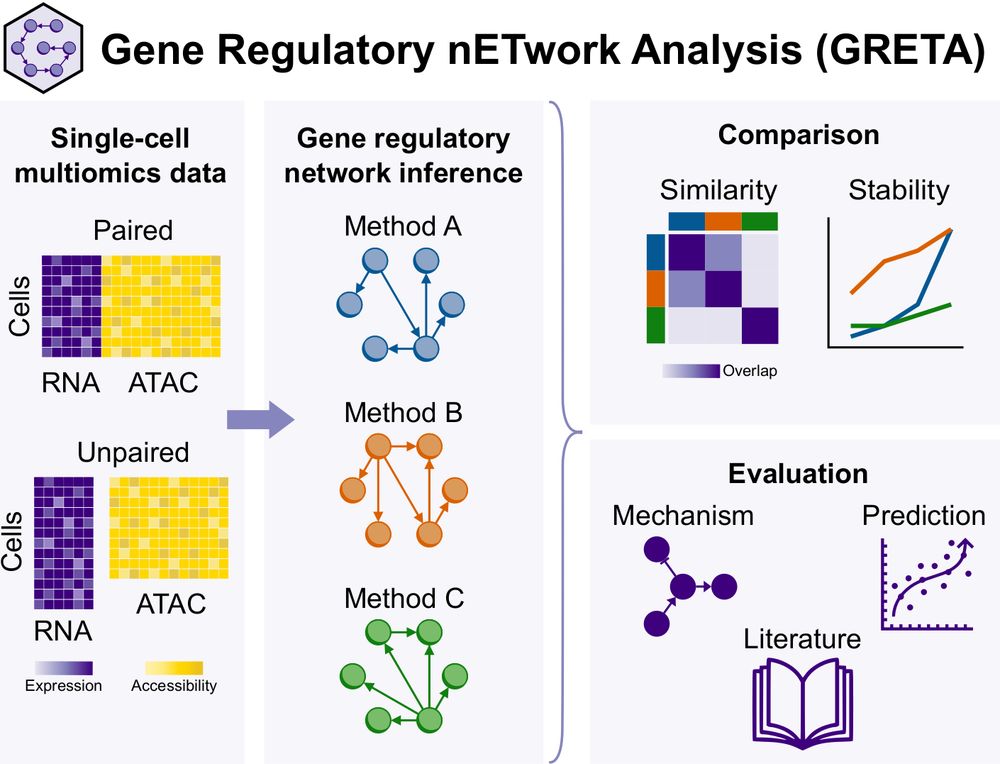
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
@steglelab.bsky.social
@steglelab.bsky.social
The increase -
• 0 years of experience = $66,300
• 1 year = $66,810
• 2 years = $67,320
🤡🤡🤡
The increase -
• 0 years of experience = $66,300
• 1 year = $66,810
• 2 years = $67,320
🤡🤡🤡
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.
@brianhiestand.bsky.social
www.science.org/doi/10.1126/...
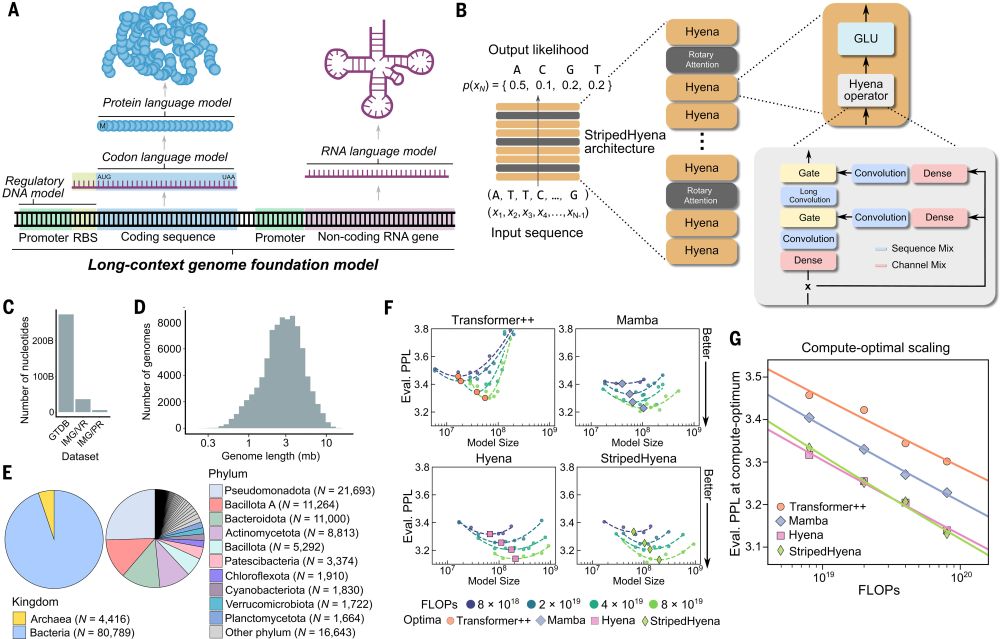
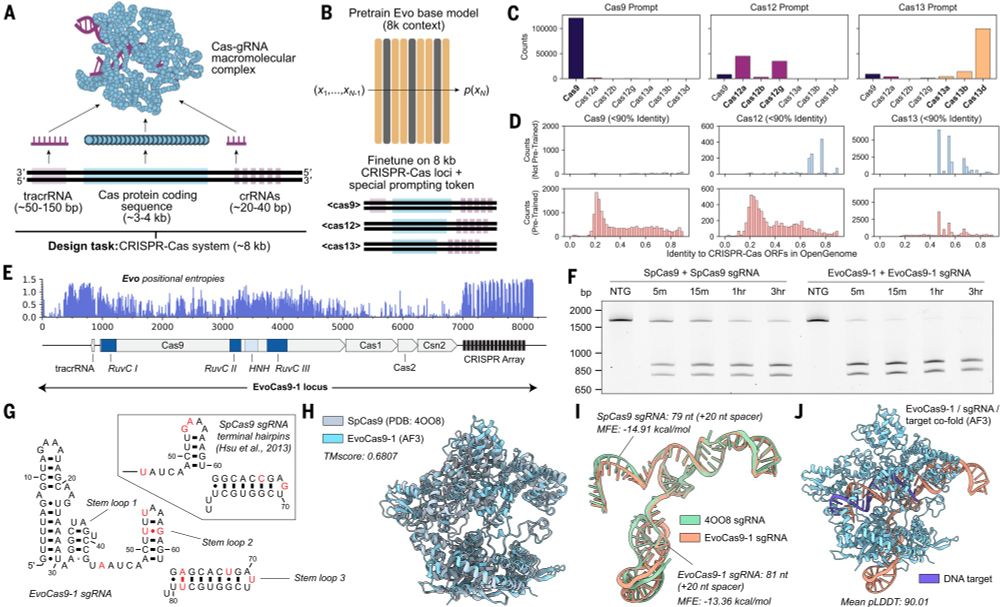
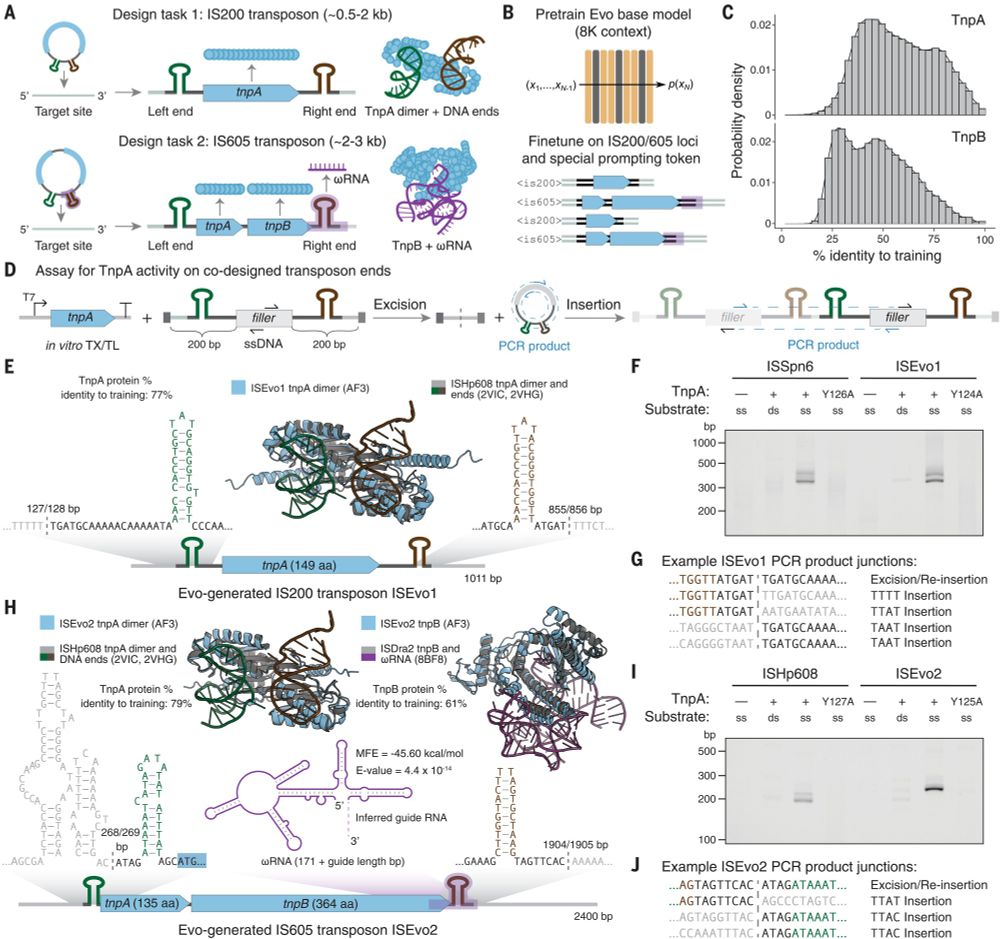
@brianhiestand.bsky.social
www.science.org/doi/10.1126/...

