www.nature.com/articles/s41...
A fun collaboration with @beckmannlab.bsky.social @doubleshuang.bsky.social

www.nature.com/articles/s41...
A fun collaboration with @beckmannlab.bsky.social @doubleshuang.bsky.social
rdcu.be/eRmJl
#ribosome #cryoEM #LMU #JHMI
rdcu.be/eRmJl
#ribosome #cryoEM #LMU #JHMI
www.nature.com/articles/s41...

www.nature.com/articles/s41...


Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
Kuffer & Marzilli engineered conditionally stable MS2 & PP7 coat proteins (dMCP & dPCP) that degrade unless bound to RNA, enabling ultra–low-background, single-mRNA imaging in live cells.
🔗 www.nature.com/articles/s41...
🧬 www.addgene.org/John_Ngo/
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Our new study shows that α-syn fibrils hijack the ESCRT membrane repair system, triggering a feedback loop that worsens aggregation.
You can find it at: authors.elsevier.com/sd/article/S...
Our new study shows that α-syn fibrils hijack the ESCRT membrane repair system, triggering a feedback loop that worsens aggregation.
You can find it at: authors.elsevier.com/sd/article/S...
❕Paper: www.nature.com/articles/s41...
❕Press Release: www.biochem.mpg.de/structure-of...
#Alzheimers @natsmb.nature.com @pyustecheca.bsky.social

❕Paper: www.nature.com/articles/s41...
❕Press Release: www.biochem.mpg.de/structure-of...
#Alzheimers @natsmb.nature.com @pyustecheca.bsky.social
#proteostasis #apolipoprotein
rdcu.be/ezRLv

#proteostasis #apolipoprotein
rdcu.be/ezRLv
More than 2700 human 3′UTRs are highly conserved. These 3′UTRs are essential components in mRNA templates, as their deletion decreases protein activity without changing protein abundance. Highly conserved 3′UTRs help the folding of proteins with long IDRs.
www.biorxiv.org/content/10.1...
More than 2700 human 3′UTRs are highly conserved. These 3′UTRs are essential components in mRNA templates, as their deletion decreases protein activity without changing protein abundance. Highly conserved 3′UTRs help the folding of proteins with long IDRs.
www.biorxiv.org/content/10.1...
By studying a protein that is difficult to fold, we discover fascinating new mechanisms by which the ribosome supports protein biogenesis.
www.biorxiv.org/content/10.1...
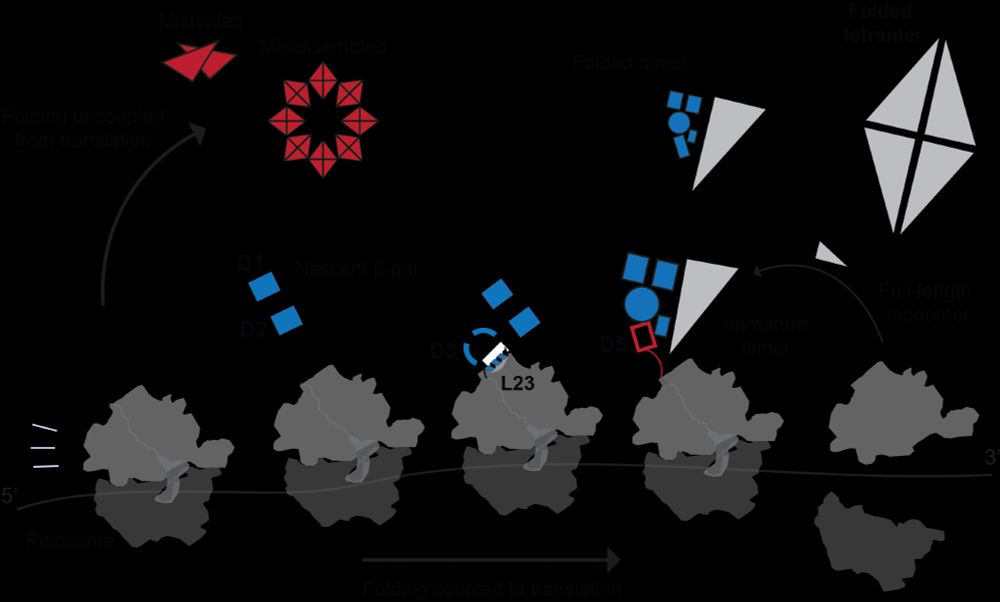
By studying a protein that is difficult to fold, we discover fascinating new mechanisms by which the ribosome supports protein biogenesis.
www.biorxiv.org/content/10.1...
Cryo-EM & functional data by Roland Beckmann and coworkers show how Bcs1 alternates between different states during its translocation activity.
www.embopress.org/doi/full/10....
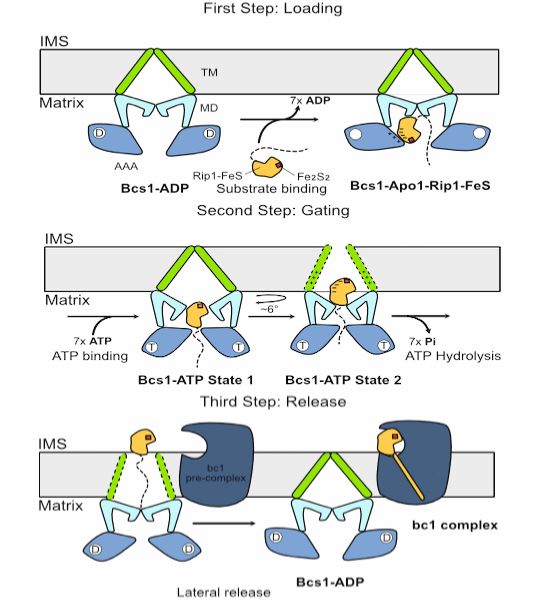
Cryo-EM & functional data by Roland Beckmann and coworkers show how Bcs1 alternates between different states during its translocation activity.
www.embopress.org/doi/full/10....

www.science.org/doi/10.1126/...
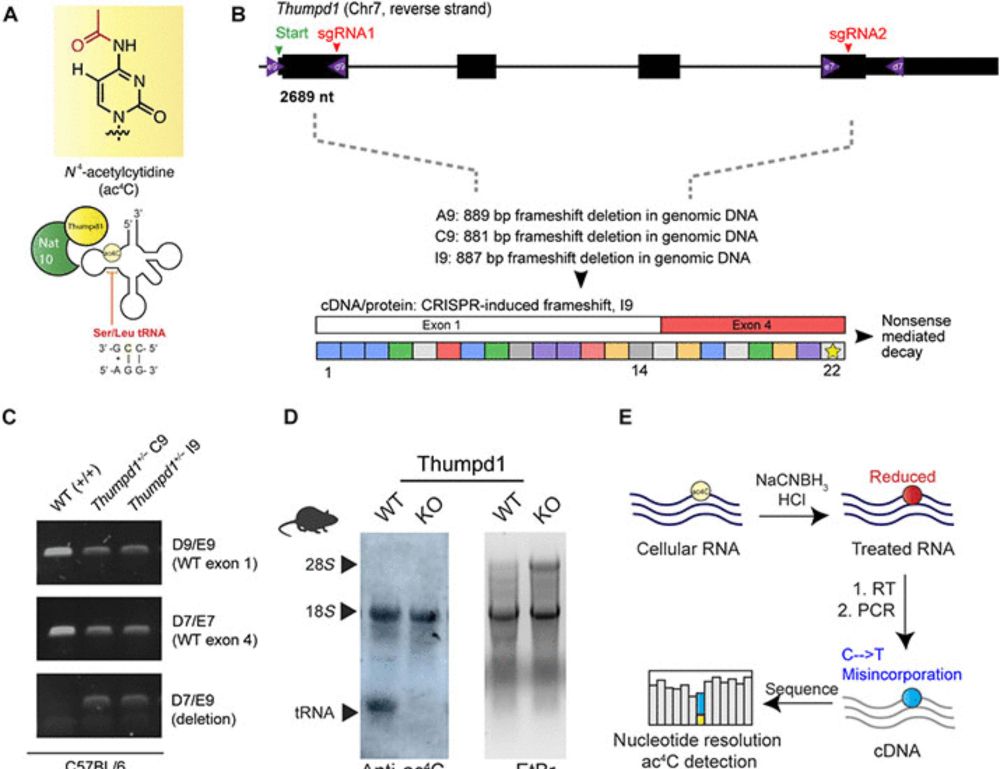
www.science.org/doi/10.1126/...
#ribosome #mRNAdecay #translation

#ribosome #mRNAdecay #translation
Dr. Farren Isaacs and his team just published in Nature:
🧬 "Engineering a genomically recoded organism with a one-stop codon"
A huge step for synthetic genomics! Read more: www.nature.com/articles/s41...

Dr. Farren Isaacs and his team just published in Nature:
🧬 "Engineering a genomically recoded organism with a one-stop codon"
A huge step for synthetic genomics! Read more: www.nature.com/articles/s41...
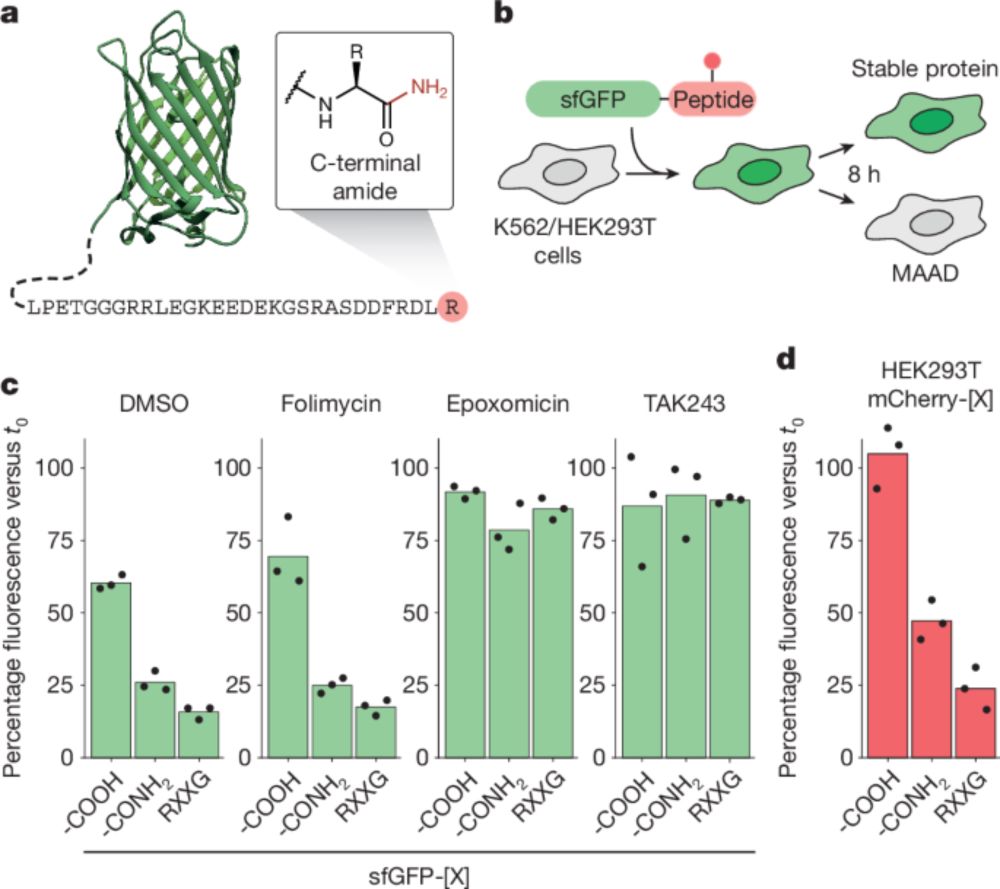
Paper: www.nature.com/articles/s41...
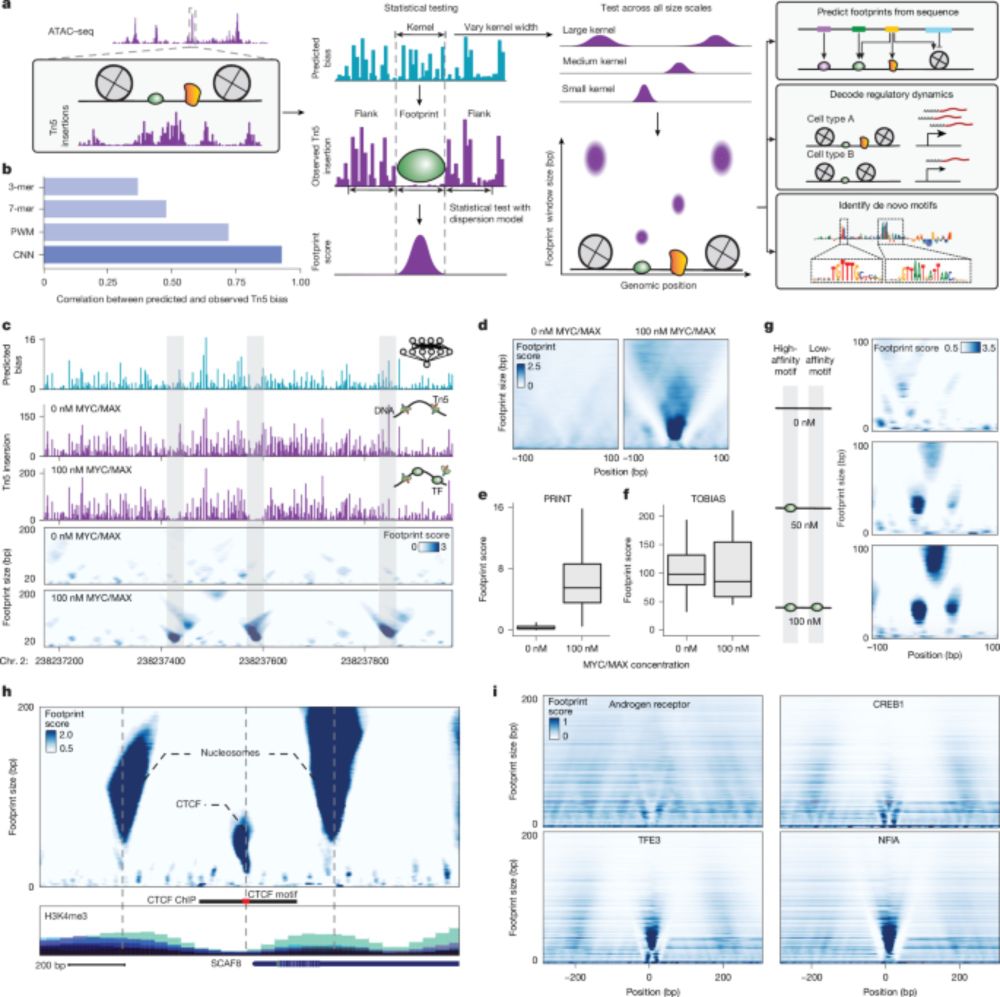
Paper: www.nature.com/articles/s41...
Learn how to make holey gold or holey carbon grids in your own laboratory!
Space is limited, so please apply at web.cvent.com/event/ee88ac...

Learn how to make holey gold or holey carbon grids in your own laboratory!
Space is limited, so please apply at web.cvent.com/event/ee88ac...
Click here to read more about it: www2.mrc-lmb.cam.ac.uk/discovery-of...
#LMBResearch
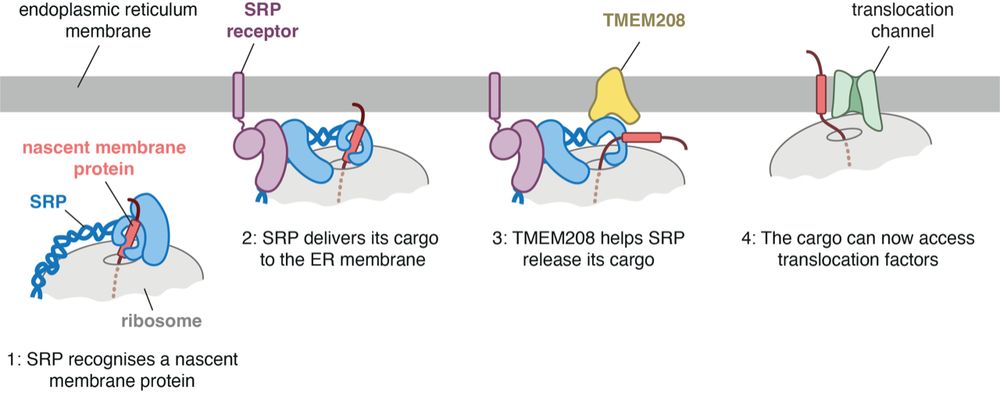
Click here to read more about it: www2.mrc-lmb.cam.ac.uk/discovery-of...
#LMBResearch
amazing work from the Hegde lab showing that this long-overlooked protein helps SRP to properly release its cargo at the ER membrane, ensuring smooth translocation. especially important for multipass membrane proteins.
www.science.org/doi/10.1126/...

amazing work from the Hegde lab showing that this long-overlooked protein helps SRP to properly release its cargo at the ER membrane, ensuring smooth translocation. especially important for multipass membrane proteins.
www.science.org/doi/10.1126/...