
@LOEWE_TBG @Senckenberg. Evolutionary genomics, gene expression, global change, novelties, climate, turtles & eels!
He/him
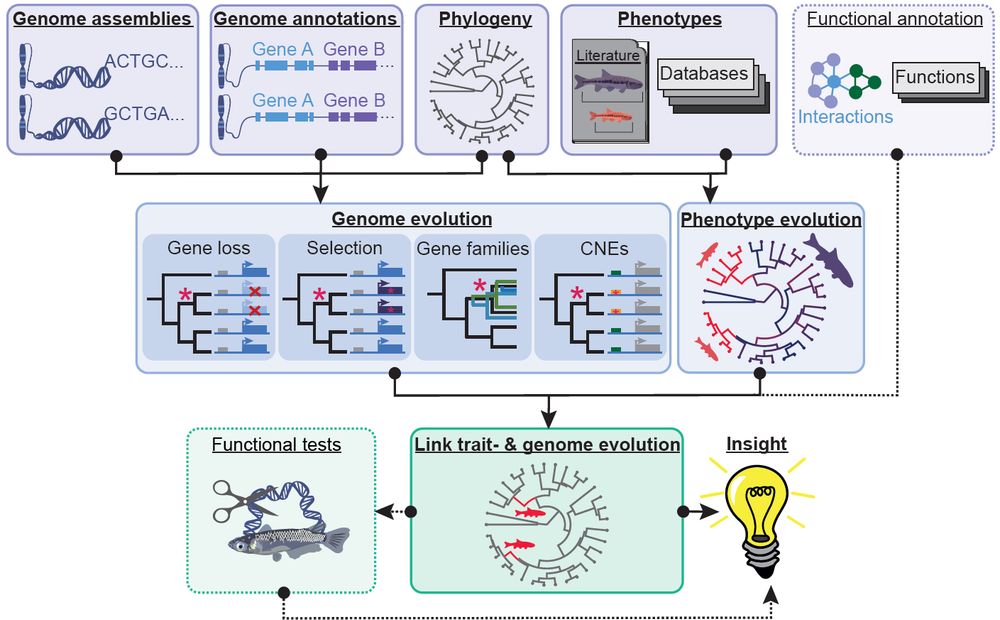
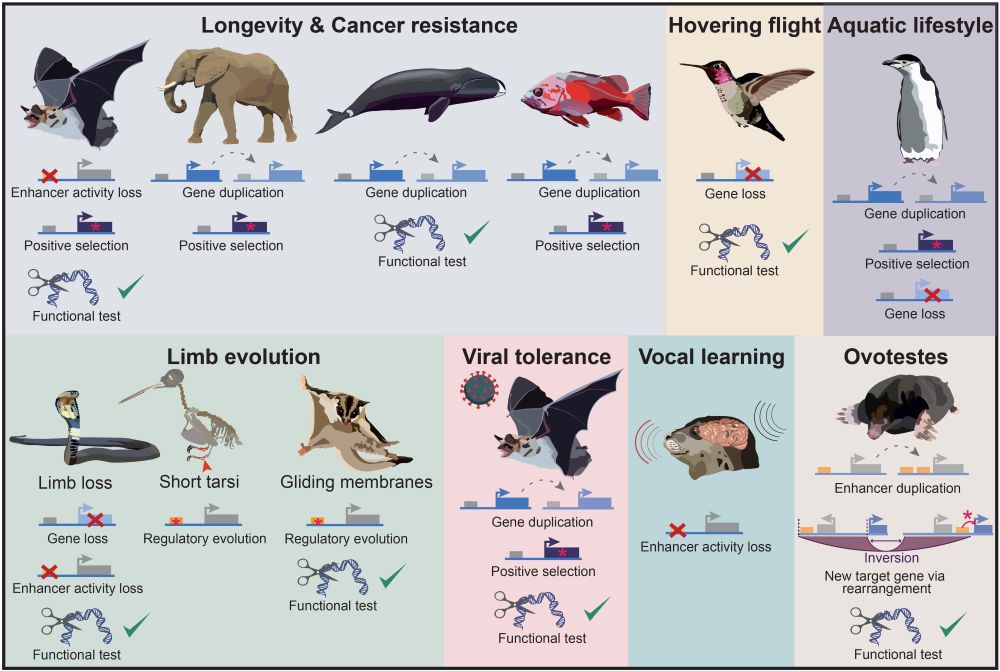
This #review is for you 📜: authors.elsevier.com/sd/article/S...
We review new #methods, remaining #challenges and #future directions and highlight recent key studies.
Thanks @hillermich.bsky.social!
Please share! 🙂
Addressing Key Challenges for High-Quality Reference Genome Generation in Europe🧬🇪🇺
Now published in Mol Ecol Res!
onlinelibrary.wiley.com/doi/10.1111/...
Thanks to the @ergabiodiv.bsky.social team!!!🙂

Addressing Key Challenges for High-Quality Reference Genome Generation in Europe🧬🇪🇺
Now published in Mol Ecol Res!
onlinelibrary.wiley.com/doi/10.1111/...
Thanks to the @ergabiodiv.bsky.social team!!!🙂
sshrc-crsh.canada.ca/en/funding/o...

sshrc-crsh.canada.ca/en/funding/o...


evol.mcmaster.ca/brian/evoldi...

evol.mcmaster.ca/brian/evoldi...
Samples are ready - you do (read: learn) everything - functional genomics, bioinformatics, genotype-phenotype associations 🤩
Please RT!
Apply by October 15th through www.jobbnorge.no/en/available...
Please share widely! 🧬🦑🖥️

Samples are ready - you do (read: learn) everything - functional genomics, bioinformatics, genotype-phenotype associations 🤩
Please RT!
The next #PopGroup meeting will take place in Lille 🍟, France, 7–9 January 2026 – just 1 hour by train from London, Brussels, and Paris.
This year, PopGroup will also host ALPHY, the annual meeting of Evolutionary Genomics.
More info: populationgeneticsgroup.org.uk
See you there !
The next #PopGroup meeting will take place in Lille 🍟, France, 7–9 January 2026 – just 1 hour by train from London, Brussels, and Paris.
This year, PopGroup will also host ALPHY, the annual meeting of Evolutionary Genomics.
More info: populationgeneticsgroup.org.uk
See you there !

Full article available: https://doi.org/10.1093/bioadv/vbaf177
Authors include: @chriswheat.bsky.social

Full article available: https://doi.org/10.1093/bioadv/vbaf177
Authors include: @chriswheat.bsky.social
This #review is for you 📜: authors.elsevier.com/sd/article/S...
We review new #methods, remaining #challenges and #future directions and highlight recent key studies.
Thanks @hillermich.bsky.social!
Please share! 🙂


This #review is for you 📜: authors.elsevier.com/sd/article/S...
We review new #methods, remaining #challenges and #future directions and highlight recent key studies.
Thanks @hillermich.bsky.social!
Please share! 🙂

Guppy males have enormous variation in color patterns, with many combinations of ornamental spots and stripes. But where does all this variation come from?

Guppy males have enormous variation in color patterns, with many combinations of ornamental spots and stripes. But where does all this variation come from?
PIs enter your position info here: forms.gle/2XTHBP6CZGEn...
Prospective students (and PIs not recruiting) share the composite list: docs.google.com/spreadsheets...
Please share! 🧪
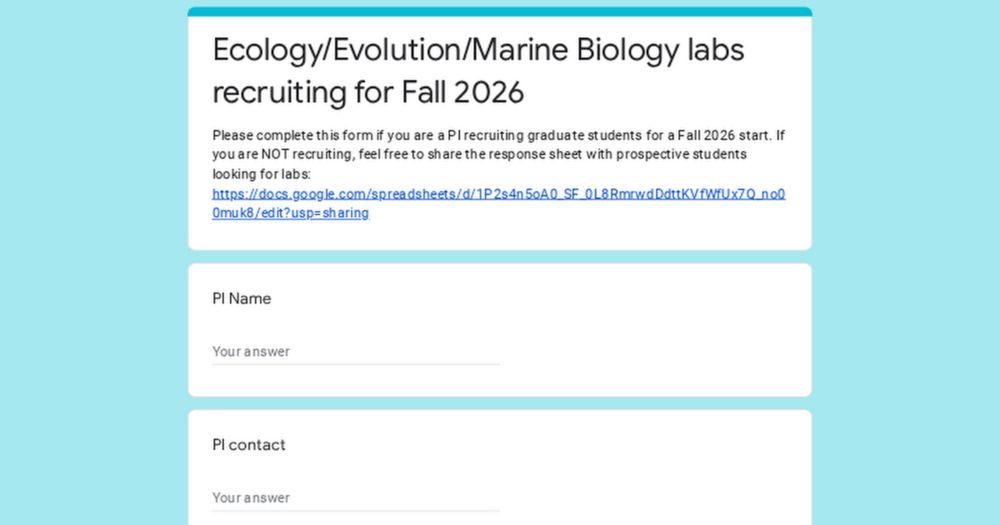
PIs enter your position info here: forms.gle/2XTHBP6CZGEn...
Prospective students (and PIs not recruiting) share the composite list: docs.google.com/spreadsheets...
Please share! 🧪
https://go.nature.com/4jLJ2C4

https://go.nature.com/4jLJ2C4
#Genomics #PopGen #Evolution #Camouflage #Birds #PhDPosition #PhDStudent
su.varbi.com/en/what:job/...

#Genomics #PopGen #Evolution #Camouflage #Birds #PhDPosition #PhDStudent
Super excited to share our new preprint “Sex decreases the pleiotropic costs of local adaptation”, where we bring a new angle to this age-old evolutionary question. Co-led by Parris Humphrey, in Michael Desai's lab. Short thread here: (1/n)
Super excited to share our new preprint “Sex decreases the pleiotropic costs of local adaptation”, where we bring a new angle to this age-old evolutionary question. Co-led by Parris Humphrey, in Michael Desai's lab. Short thread here: (1/n)
Paper: www.nature.com/articles/s41...

Paper: www.nature.com/articles/s41...
Through an unprecedented synthesis (2133 studies!) we show that humans are not only shrinking species numbers—but reshaping entire communities across the planet. 🌍🌐🐟🌿🪲
www.nature.com/articles/s41...
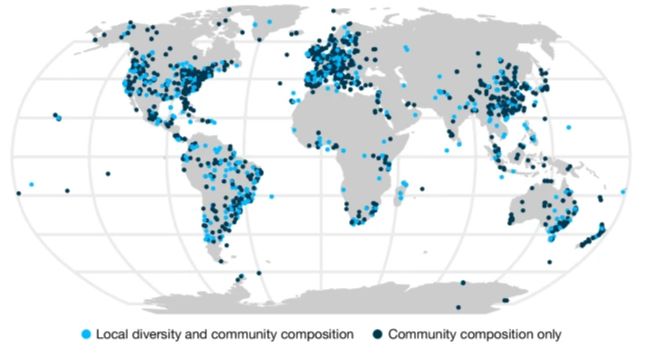
Through an unprecedented synthesis (2133 studies!) we show that humans are not only shrinking species numbers—but reshaping entire communities across the planet. 🌍🌐🐟🌿🪲
www.nature.com/articles/s41...
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
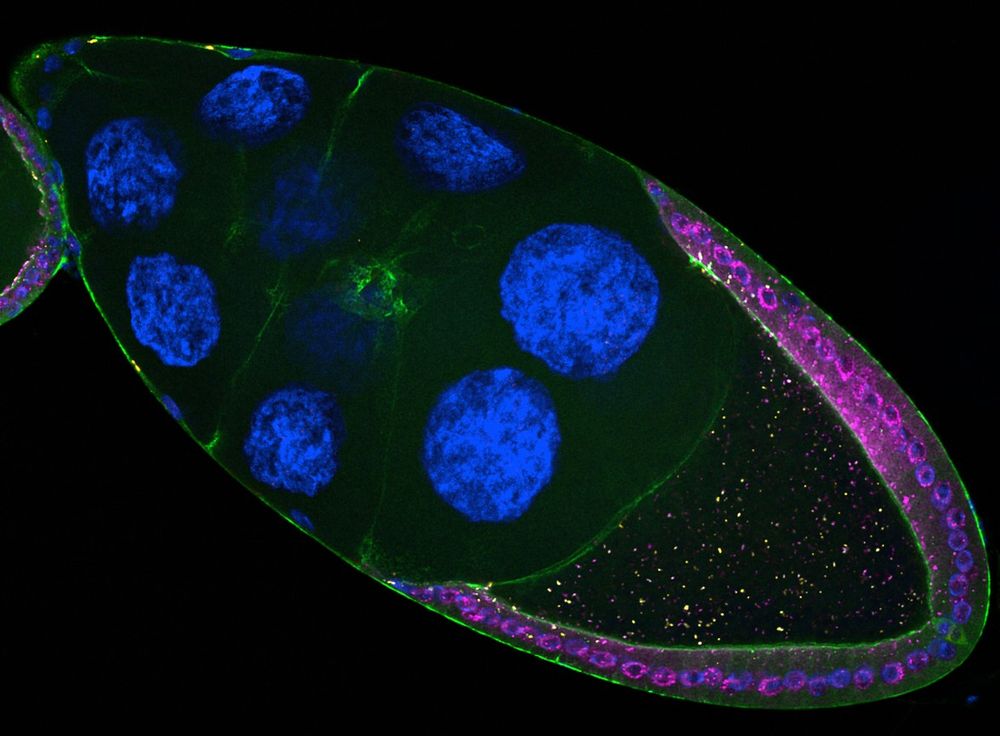
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
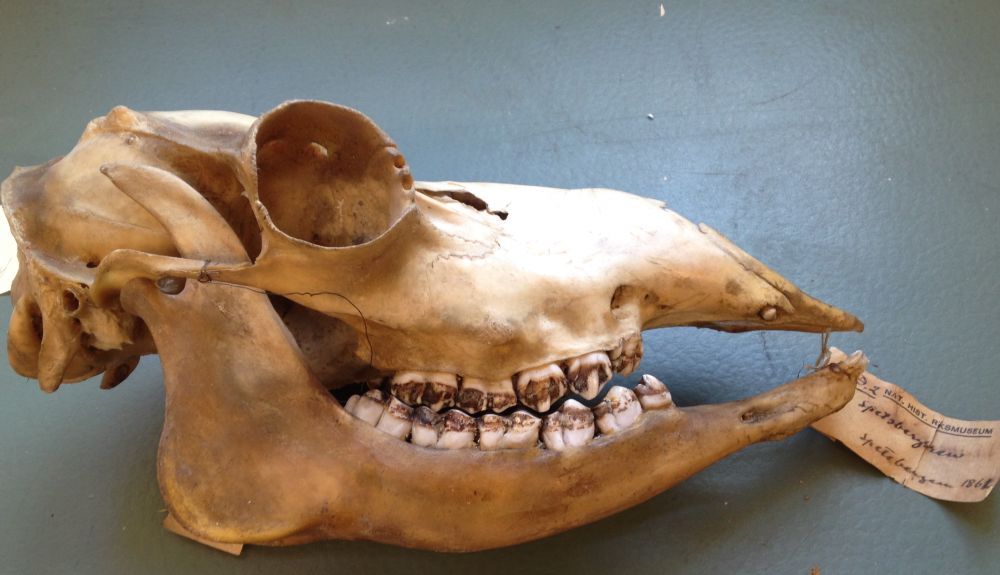
genomebiology.biomedcentral.com/articles/10....
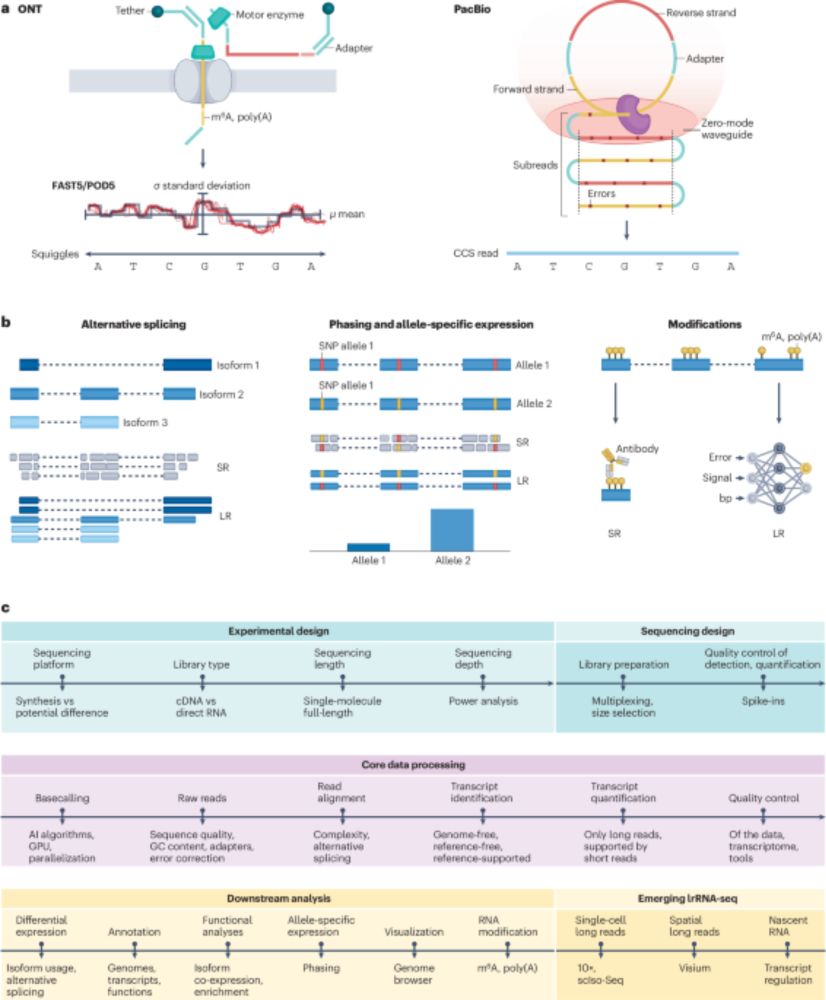
We show that specimens that have been stored in ethanol for decades can still be amenable for long-read sequencing:🧵
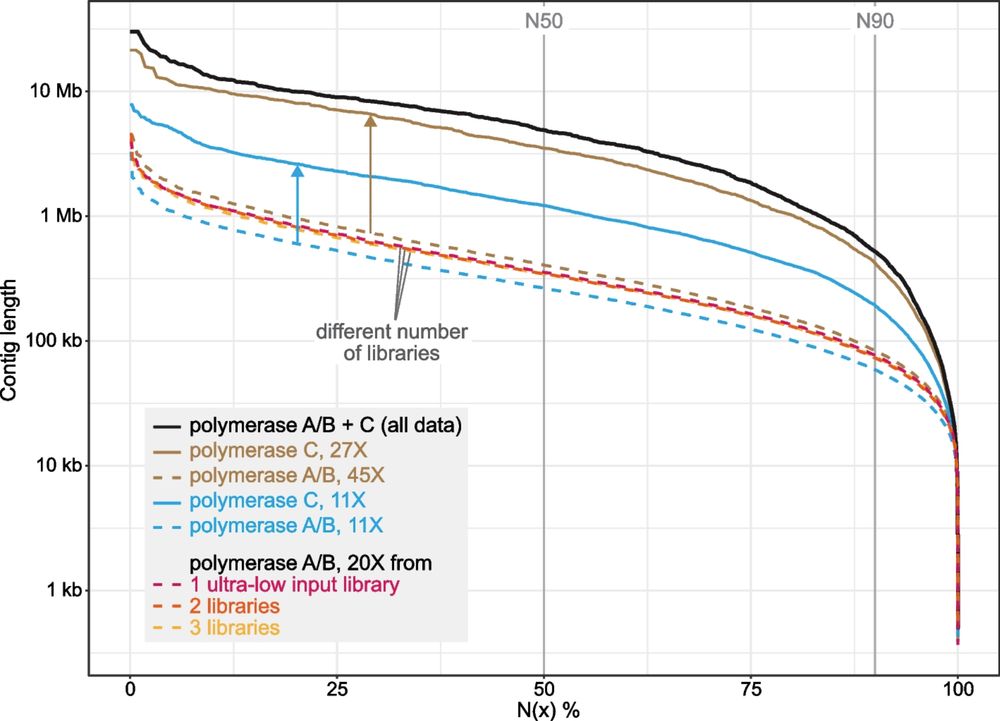
genomebiology.biomedcentral.com/articles/10....
We show that specimens that have been stored in ethanol for decades can still be amenable for long-read sequencing:🧵

