genomebiology.biomedcentral.com/articles/10....
We show that specimens that have been stored in ethanol for decades can still be amenable for long-read sequencing:🧵
Keynote #StefanMundlos: Functional Genomics of Bat Limb Development
With Genome Chalk team talks #LucasCanesin, #YuryMalovichko, #AlejandroGonzales, #BernhardBein & #MicheleAlbertini

Keynote #StefanMundlos: Functional Genomics of Bat Limb Development
With Genome Chalk team talks #LucasCanesin, #YuryMalovichko, #AlejandroGonzales, #BernhardBein & #MicheleAlbertini
🔗 Release notes and source code: github.com/vgl-hub/rdev...
🔗 Release notes and source code: github.com/vgl-hub/rdev...

https://www.biorxiv.org/content/10.1101/2025.11.18.689136v1
Several lineages of #birds and #mammals appear to have independently gained or lost the ability to enter #torpor. 🧵1/4
https://www.biorxiv.org/content/10.1101/2025.11.18.689136v1
Several lineages of #birds and #mammals appear to have independently gained or lost the ability to enter #torpor. 🧵1/4
We produced complete genomes for 2 Xenarthra and placed them in a mammalian comparative framework. We found that Xenarthra harbour the largest number of retrocopies in mammals! www.biorxiv.org/content/10.1...

We produced complete genomes for 2 Xenarthra and placed them in a mammalian comparative framework. We found that Xenarthra harbour the largest number of retrocopies in mammals! www.biorxiv.org/content/10.1...

Key finding: Some CRISPR-mutations may only emerge in the second generation, due to germ cell mosaicism
www.biorxiv.org/content/10.1...

Key finding: Some CRISPR-mutations may only emerge in the second generation, due to germ cell mosaicism
www.biorxiv.org/content/10.1...
go.nature.com/4mOb5T9

go.nature.com/4mOb5T9
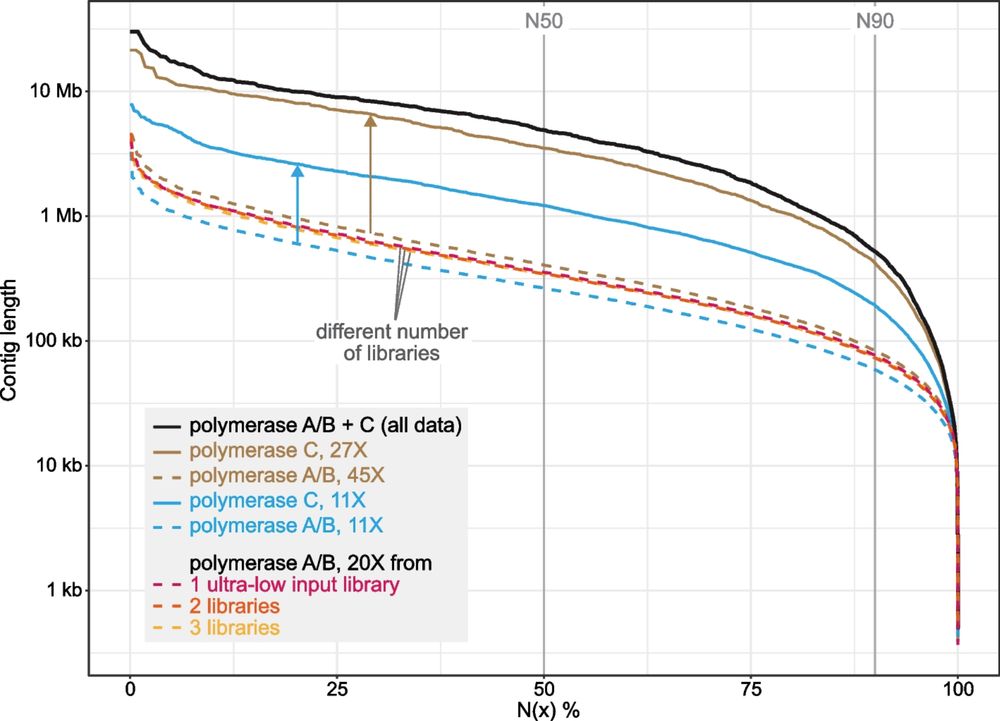
Really cool paper showing that Ultra-low input (ULI) HiFi sequencing can help cancer diagnostics.
Only emphasizes how interdisciplinary the ULI field is ( #museomics, #single-cell #sequencing, #diagnostics etc)
www.medrxiv.org/content/10.1...

Really cool paper showing that Ultra-low input (ULI) HiFi sequencing can help cancer diagnostics.
Only emphasizes how interdisciplinary the ULI field is ( #museomics, #single-cell #sequencing, #diagnostics etc)
www.medrxiv.org/content/10.1...

💻️ rdeval code: https://github.com/vgl-hub/rdeval (3/3)

💻️ rdeval code: https://github.com/vgl-hub/rdeval (3/3)
This #review is for you 📜: authors.elsevier.com/sd/article/S...
We review new #methods, remaining #challenges and #future directions and highlight recent key studies.
Thanks @hillermich.bsky.social!
Please share! 🙂
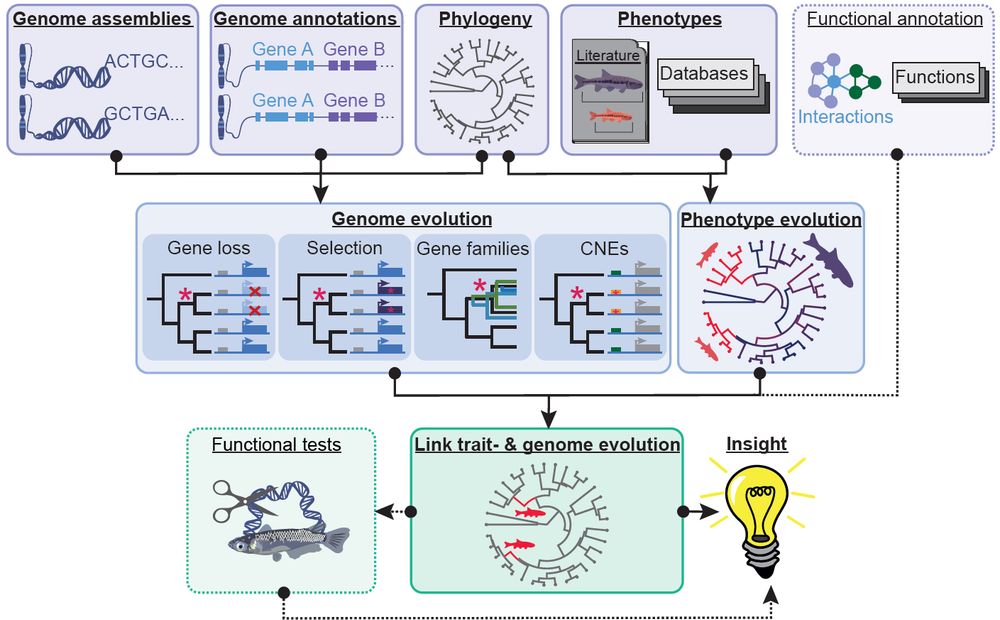
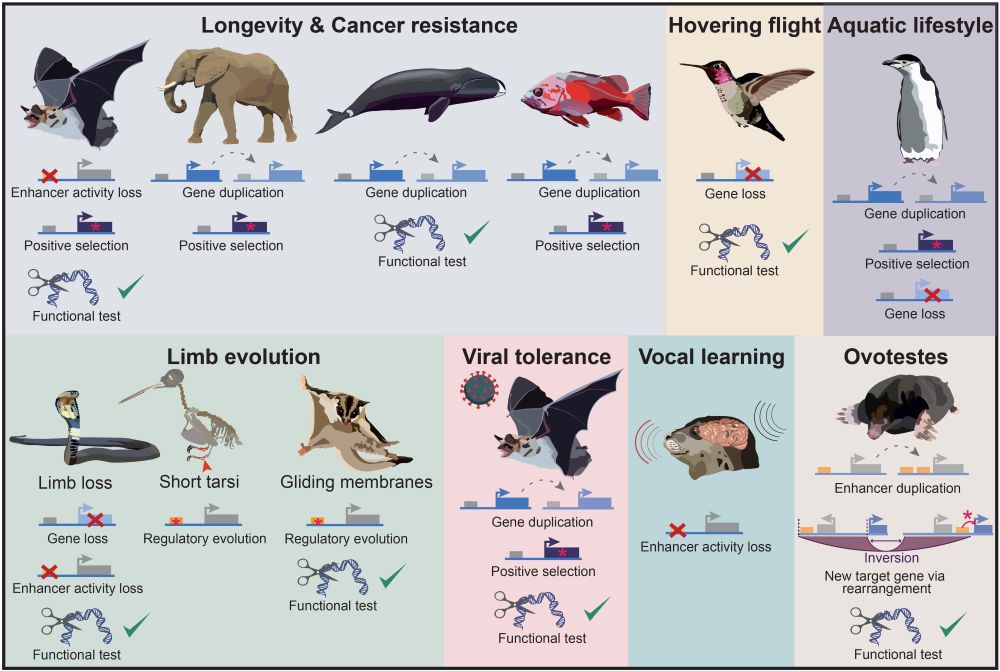
This #review is for you 📜: authors.elsevier.com/sd/article/S...
We review new #methods, remaining #challenges and #future directions and highlight recent key studies.
Thanks @hillermich.bsky.social!
Please share! 🙂
Congrats!!
zookeys.pensoft.net/articles.php...
#FreshwaterCrab #Biodiversity #Potamidae

Congrats!!
zookeys.pensoft.net/articles.php...
#FreshwaterCrab #Biodiversity #Potamidae

#PCRAmplification #LongReadSequencing
Now, we’re sharing how this protocol is helping bring #HiFisequencing to FFPE samples in cancer research.
🔗 Read the update: bit.ly/4dkvkDO
#PacBio #CancerResearch

#PCRAmplification #LongReadSequencing
🔗 rdcu.be/eiHRG
@sgn.one @leibnizlib.bsky.social #biodiversity #genomics

🔗 rdcu.be/eiHRG
@sgn.one @leibnizlib.bsky.social #biodiversity #genomics
1. MDA on pg input DNA for ONT rapid seq of pathogens + Tool to detect concatemers
doi.org/10.1111/1755...
2. MDA for metagenomics of low biomass samples
doi.org/10.1093/isme...
Exciting Stuff, small inputs + ampli. going strong!

1. MDA on pg input DNA for ONT rapid seq of pathogens + Tool to detect concatemers
doi.org/10.1111/1755...
2. MDA for metagenomics of low biomass samples
doi.org/10.1093/isme...
Exciting Stuff, small inputs + ampli. going strong!
5 beeindruckende Weichtiere stehen im Finale. Dem Gewinner-Weichtier winkt die Entschlüsselung seines kompletten Genoms! 🧬🏆
Erfahrt mehr über die Kandidaten und stimmt bis zum 31.3. für euren Favoriten ab 👉 moty.senckenberg.science



5 beeindruckende Weichtiere stehen im Finale. Dem Gewinner-Weichtier winkt die Entschlüsselung seines kompletten Genoms! 🧬🏆
Erfahrt mehr über die Kandidaten und stimmt bis zum 31.3. für euren Favoriten ab 👉 moty.senckenberg.science
@sgn.one @sigwartae.bsky.social @kmkocot.bsky.social @squamiferum.bsky.social @vans-gonzalez.bsky.social @cgreve.bsky.social @mkyapchiongco.bsky.social
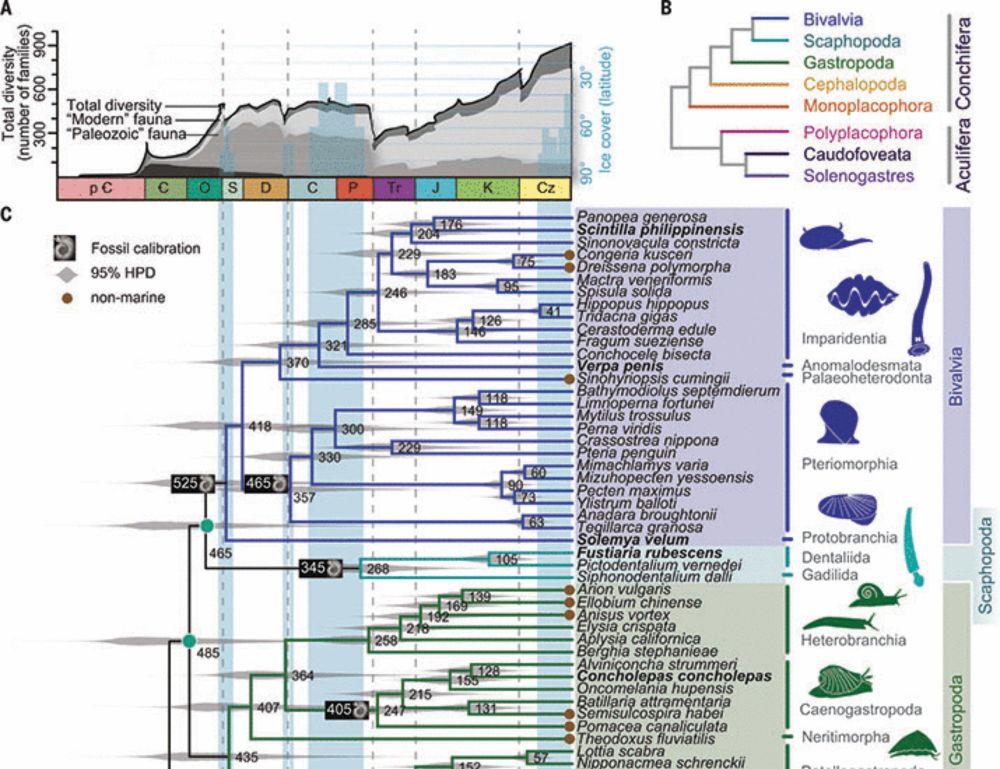
@sgn.one @sigwartae.bsky.social @kmkocot.bsky.social @squamiferum.bsky.social @vans-gonzalez.bsky.social @cgreve.bsky.social @mkyapchiongco.bsky.social
genomebiology.biomedcentral.com/articles/10....
#PacBio
#HiFiSequencing
#PCR
genomebiology.biomedcentral.com/articles/10....
#PacBio
#HiFiSequencing
#PCR
genome.cshlp.org/content/35/2...

genome.cshlp.org/content/35/2...
@yanischrysost.bsky.social Larissa Arantes, Tom Brown, Charlotte Gerheim, Tilman Schell !!!
@yanischrysost.bsky.social Larissa Arantes, Tom Brown, Charlotte Gerheim, Tilman Schell !!!

