The assemblies 3.1 Gb size also surpasses the previous 500 Mb protocol size limit.
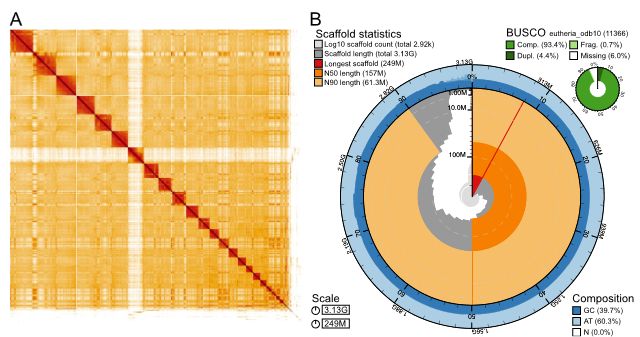
The assemblies 3.1 Gb size also surpasses the previous 500 Mb protocol size limit.
Aligning reads of B. torquatus to another sloth species, we saw obvious read drop-outs that polymerase C reads could cover:
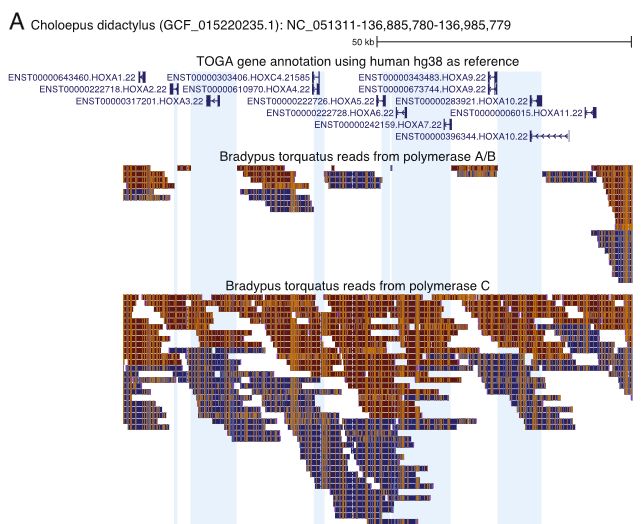
Aligning reads of B. torquatus to another sloth species, we saw obvious read drop-outs that polymerase C reads could cover:
This way, we could sequence the maned sloth Bradypus torquatus to a coverage of 45X.
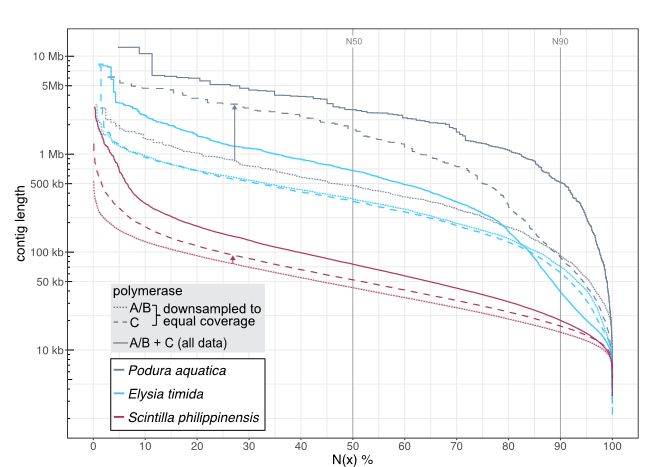
However, contig N50 of the assembly was below 1 Mb:
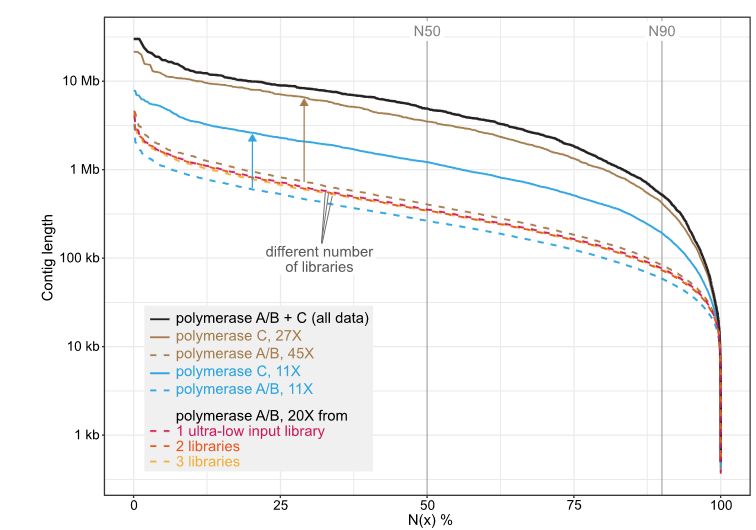
This way, we could sequence the maned sloth Bradypus torquatus to a coverage of 45X.
However, contig N50 of the assembly was below 1 Mb:

