pubs.acs.org/doi/full/10....
pubs.acs.org/doi/full/10....
Sciex - New ZenoTOF 8600, Echo-DMS
Thermo - New Exploris, New Astral (one of these is named Excedion), New OptiFlow source
Agilent - New single quad
Bruker - OmniTIMS, new triple quad, new LC
Sciex - New ZenoTOF 8600, Echo-DMS
Thermo - New Exploris, New Astral (one of these is named Excedion), New OptiFlow source
Agilent - New single quad
Bruker - OmniTIMS, new triple quad, new LC
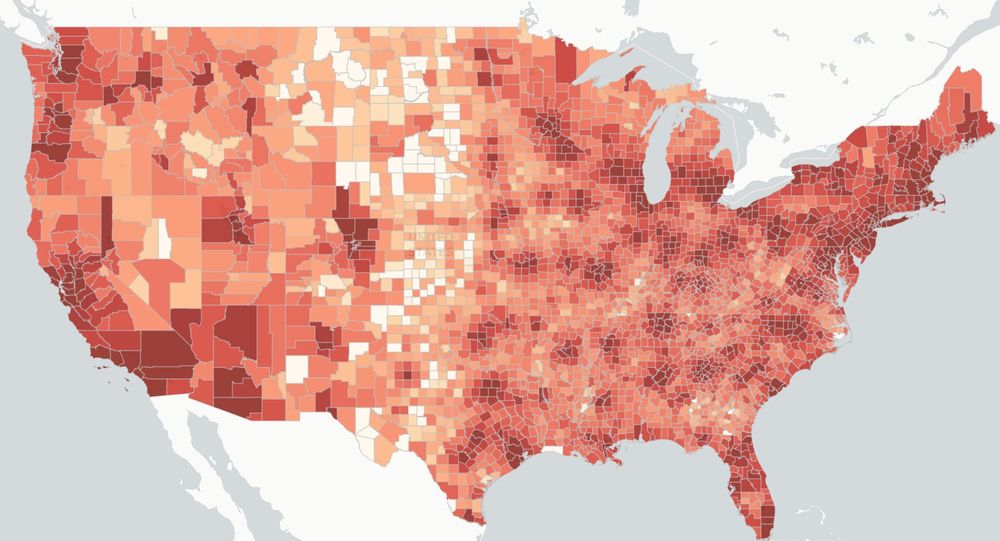
Find out how your community may be impacted.
Explore more at SCIMaP: scienceimpacts.org
a 🧵
Find out how your community may be impacted.
Explore more at SCIMaP: scienceimpacts.org
a 🧵



This helps me remember what this is all about.
Ironically, it is about the treatment of pain.

This helps me remember what this is all about.
Ironically, it is about the treatment of pain.
---
#proteomics #prot-other

---
#proteomics #prot-other
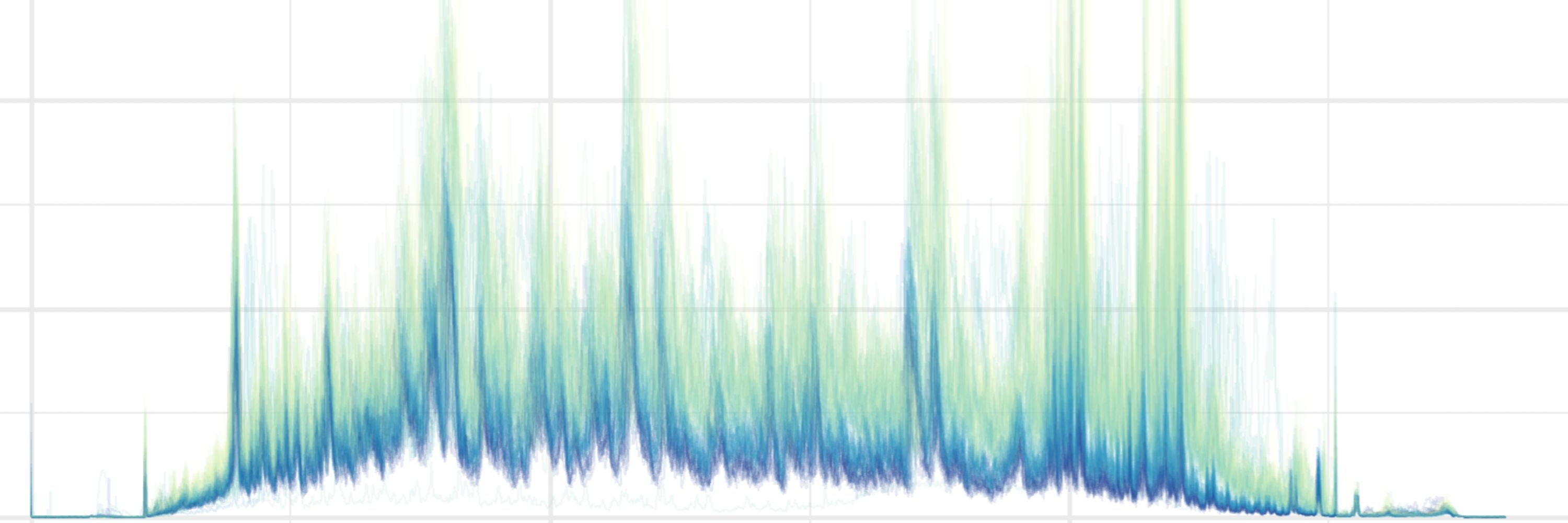
Weather’s going wild, and now your mass spec data’s a mess too?
Coindidence? Nope!
We reveal how weather-driven air pressure fluctuations impact diaPASEF-based high troughput proteomics - and how to fix it!
Check out our new paper! #diaPASEF #Weatheromics
pubs.acs.org/doi/10.1021/...

Weather’s going wild, and now your mass spec data’s a mess too?
Coindidence? Nope!
We reveal how weather-driven air pressure fluctuations impact diaPASEF-based high troughput proteomics - and how to fix it!
Check out our new paper! #diaPASEF #Weatheromics
pubs.acs.org/doi/10.1021/...

www.mcponline.org/article/S153...
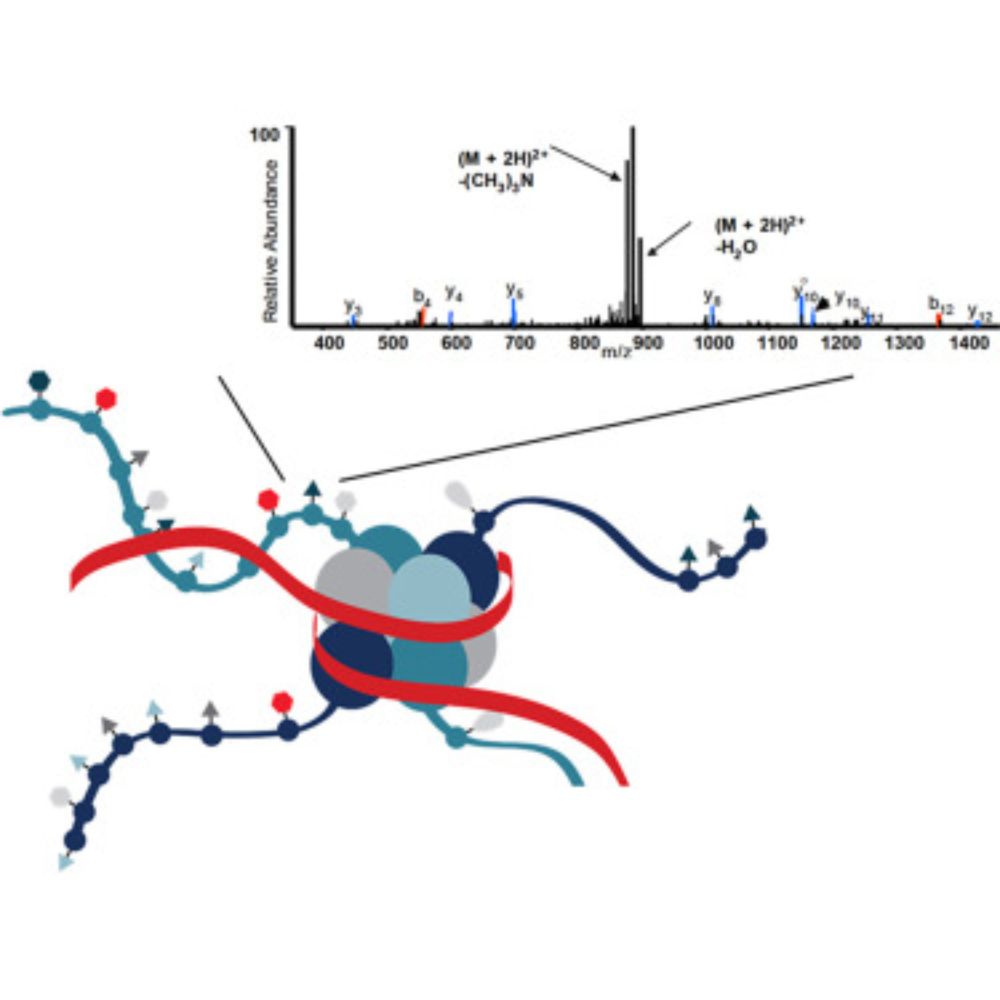
www.mcponline.org/article/S153...
www.mcponline.org/article/S153...
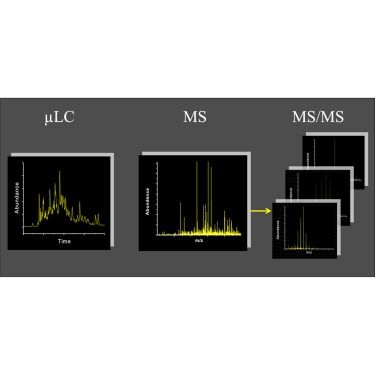
www.mcponline.org/article/S153...


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
communities.springernature.com/posts/wrangl...
communities.springernature.com/posts/wrangl...
2.8 million high-confidence peptide-spectrum matches derived from nine different species from wnoble.bsky.social for #teammassspec

2.8 million high-confidence peptide-spectrum matches derived from nine different species from wnoble.bsky.social for #teammassspec

