🗓️ 20 Nov 2025
📍 Human Technopole, Milan
Join us for a one-day dive into:
🔹 Cancer genomics
🔹 CRISPR screening
🔹 AI-driven target discovery
With the patronage of AIRC
🔗 humantechnopole.it/en/trainings...

🗓️ 20 Nov 2025
📍 Human Technopole, Milan
Join us for a one-day dive into:
🔹 Cancer genomics
🔹 CRISPR screening
🔹 AI-driven target discovery
With the patronage of AIRC
🔗 humantechnopole.it/en/trainings...
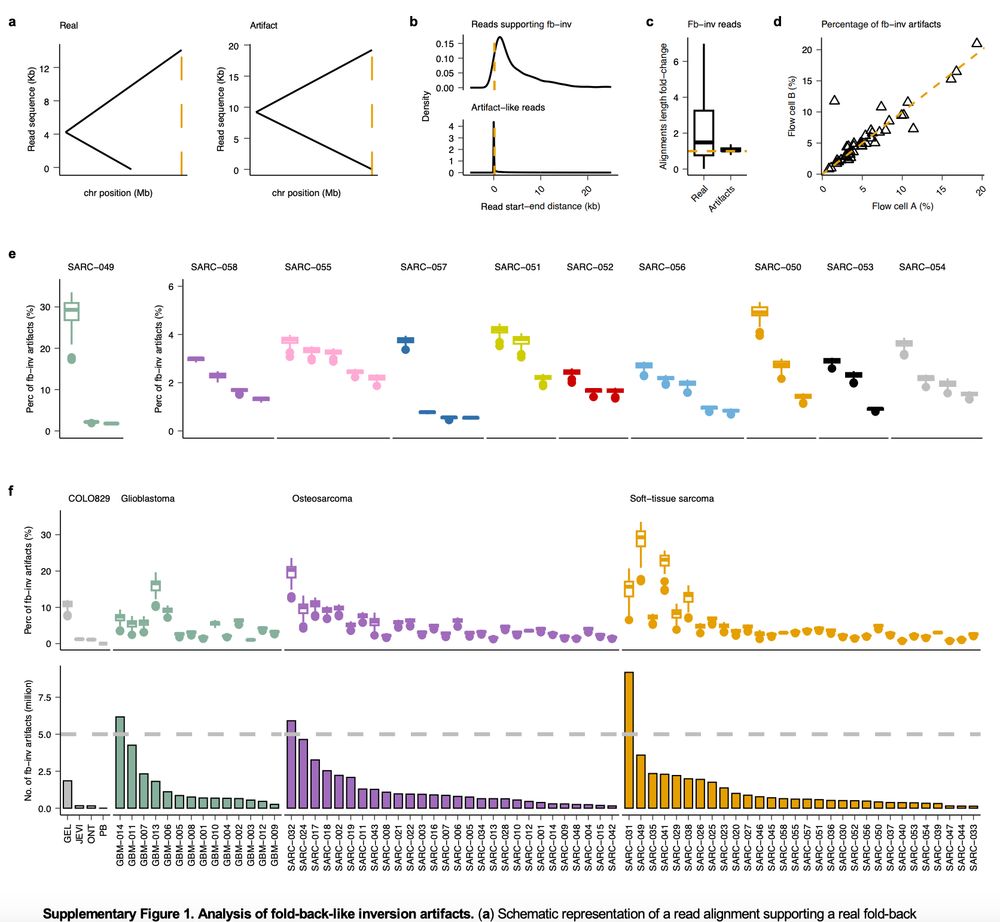
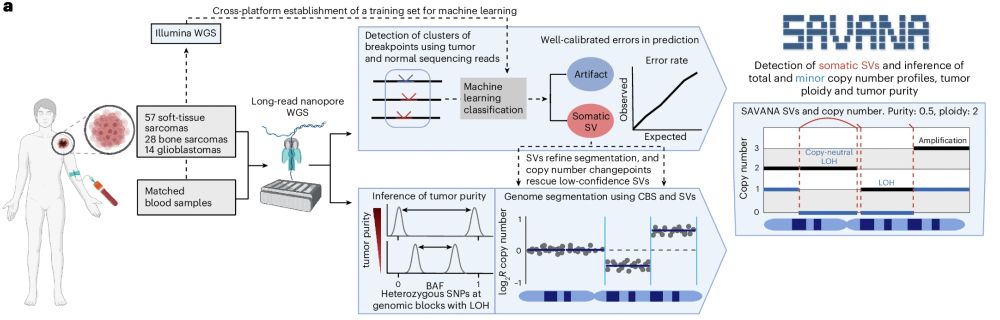

Read about it here 👇
https://bit.ly/4kyM7...
Read about it here 👇
https://bit.ly/4kyM7...
We are a mission-driven, highly collaborative and dynamic team located at the Wellcome Genome Campus, one of the most exciting hubs in the world focused on biomedical research 👇

We are a mission-driven, highly collaborative and dynamic team located at the Wellcome Genome Campus, one of the most exciting hubs in the world focused on biomedical research 👇
If you're interested in #ecDNA and want to hear or present groundbreaking research, register now. Abstract submission deadline is 14 March.
Learn more: www.cancergrandchallenges.org/ecDNA-2025

If you're interested in #ecDNA and want to hear or present groundbreaking research, register now. Abstract submission deadline is 14 March.
Learn more: www.cancergrandchallenges.org/ecDNA-2025
doi.org/10.1158/2159...

doi.org/10.1158/2159...

