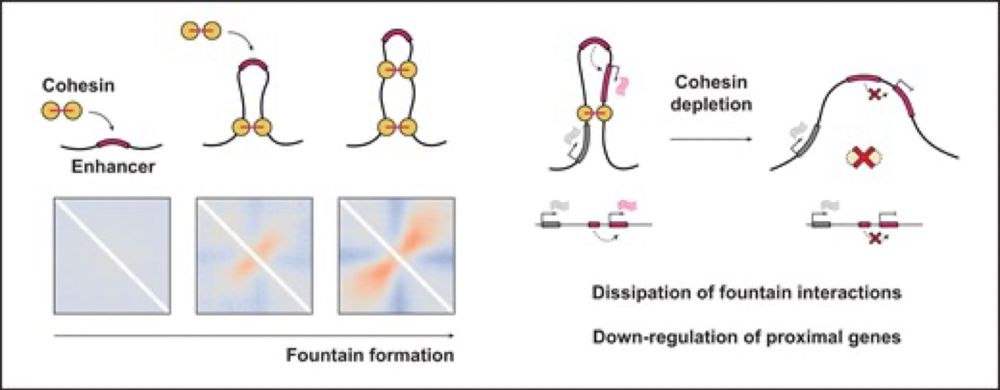
A short thread follows 1/n

A short thread follows 1/n
@jbuenrostro.bsky.social @eboyden3.bsky.social + al @broadinstitute.org @harvard.edu
#ExIGS #Chromatin #Lamin #Progeria #Aging #ExpansionMicroscopy
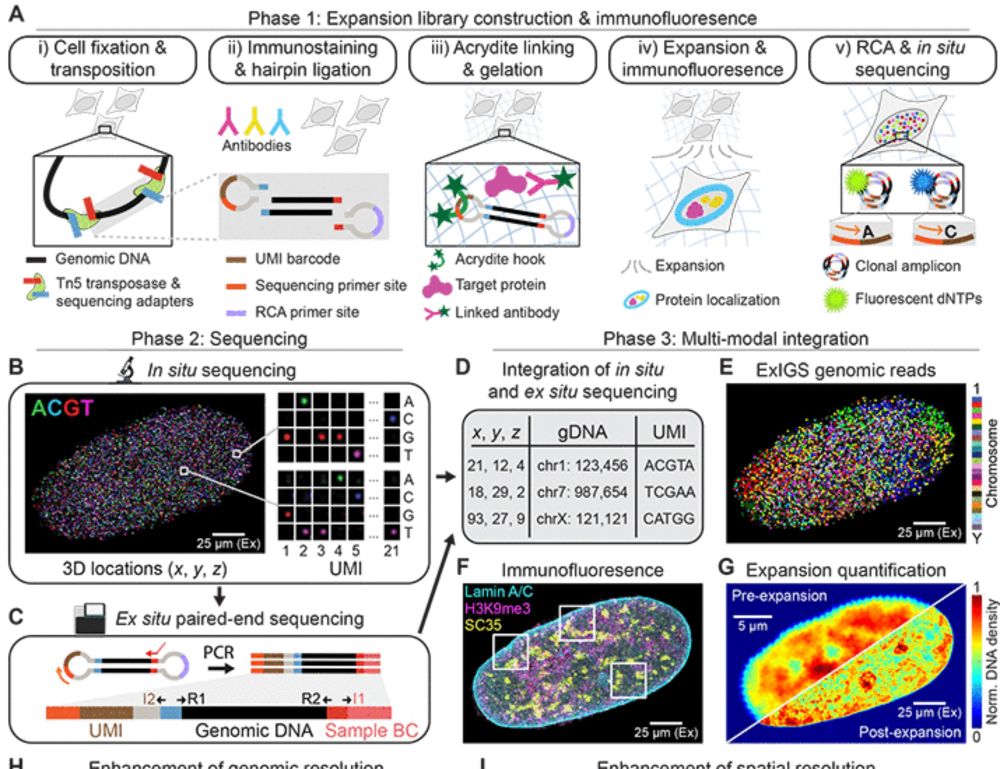
@jbuenrostro.bsky.social @eboyden3.bsky.social + al @broadinstitute.org @harvard.edu
#ExIGS #Chromatin #Lamin #Progeria #Aging #ExpansionMicroscopy
pubmed.ncbi.nlm.nih.gov/40436628/
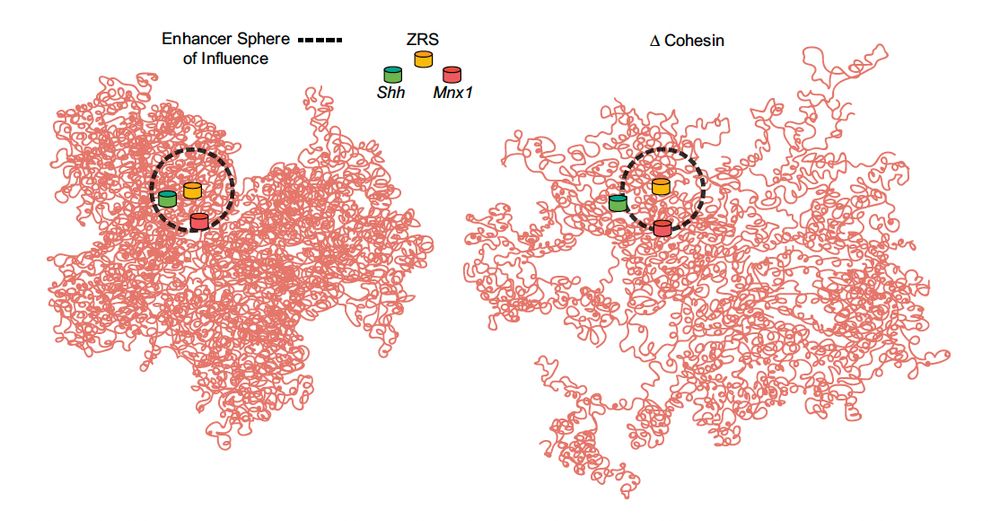
pubmed.ncbi.nlm.nih.gov/40436628/
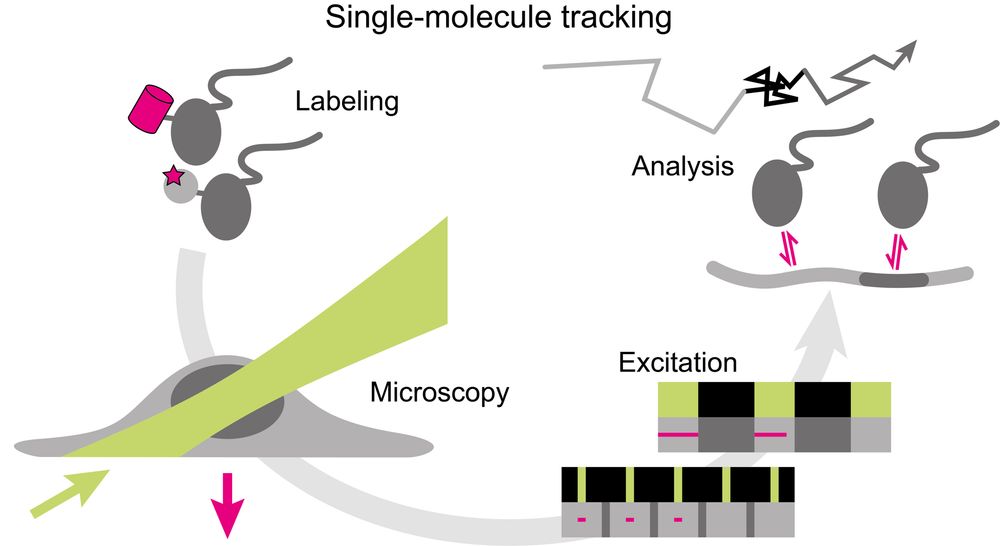
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
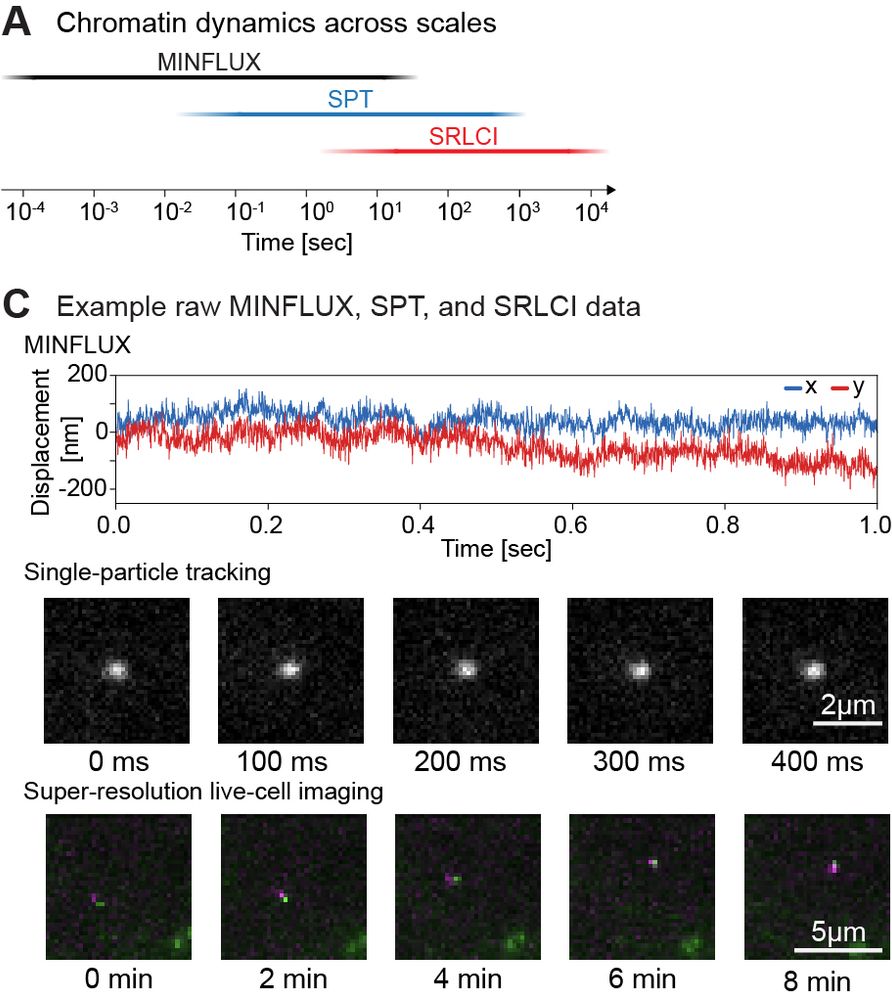
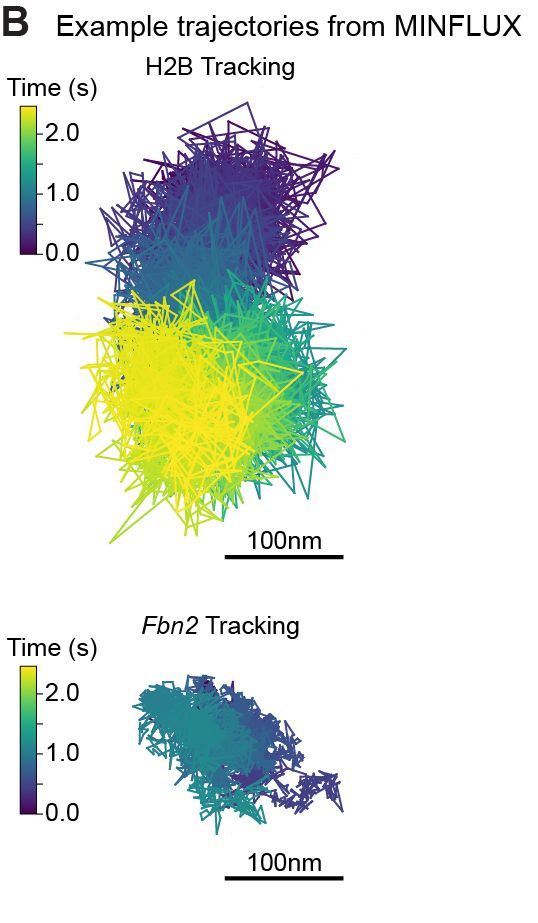
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Through these twin controls,
cells protect their proteins well,
even through shifts.
Thus, life adapts fast—
a story of balance told
through turnover rates.
🥳 Fantastic work spearheaded by Michael Sun and Benjamin Martin
www.biorxiv.org/content/10.1...

Registration is open! 👇
www.embl.org/about/info/c...

Registration is open! 👇
www.embl.org/about/info/c...
doi.org/10.1038/s414...
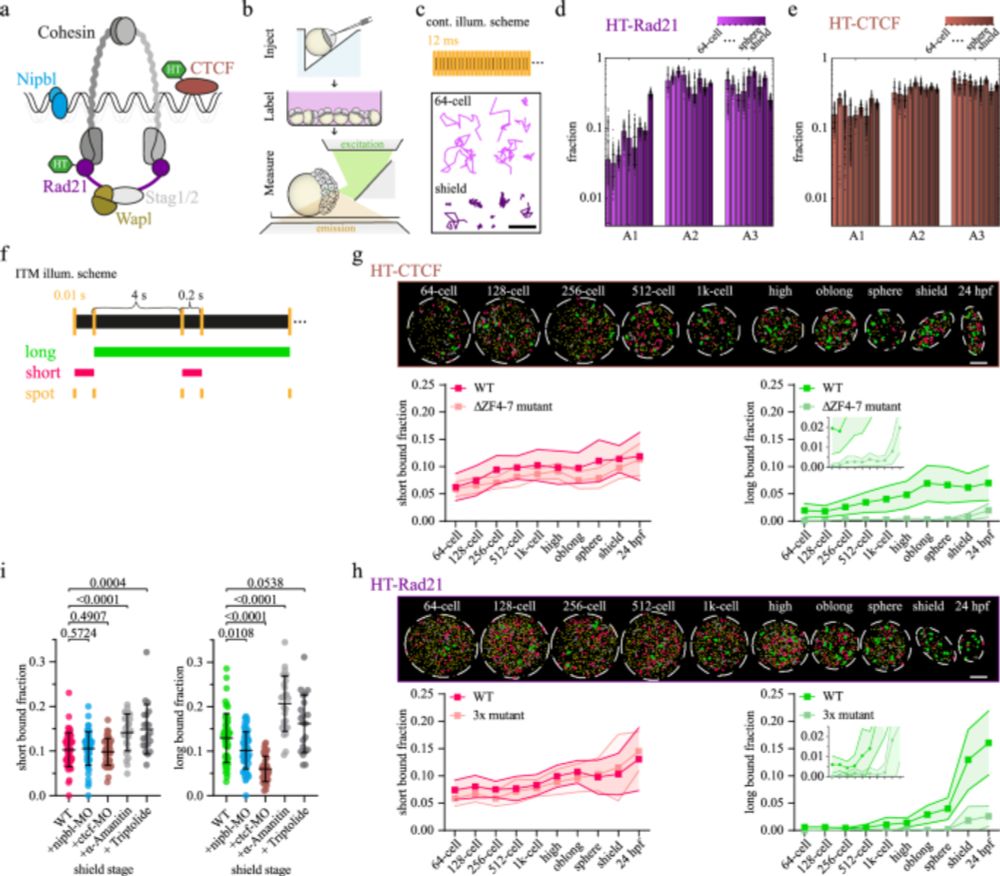
doi.org/10.1038/s414...
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
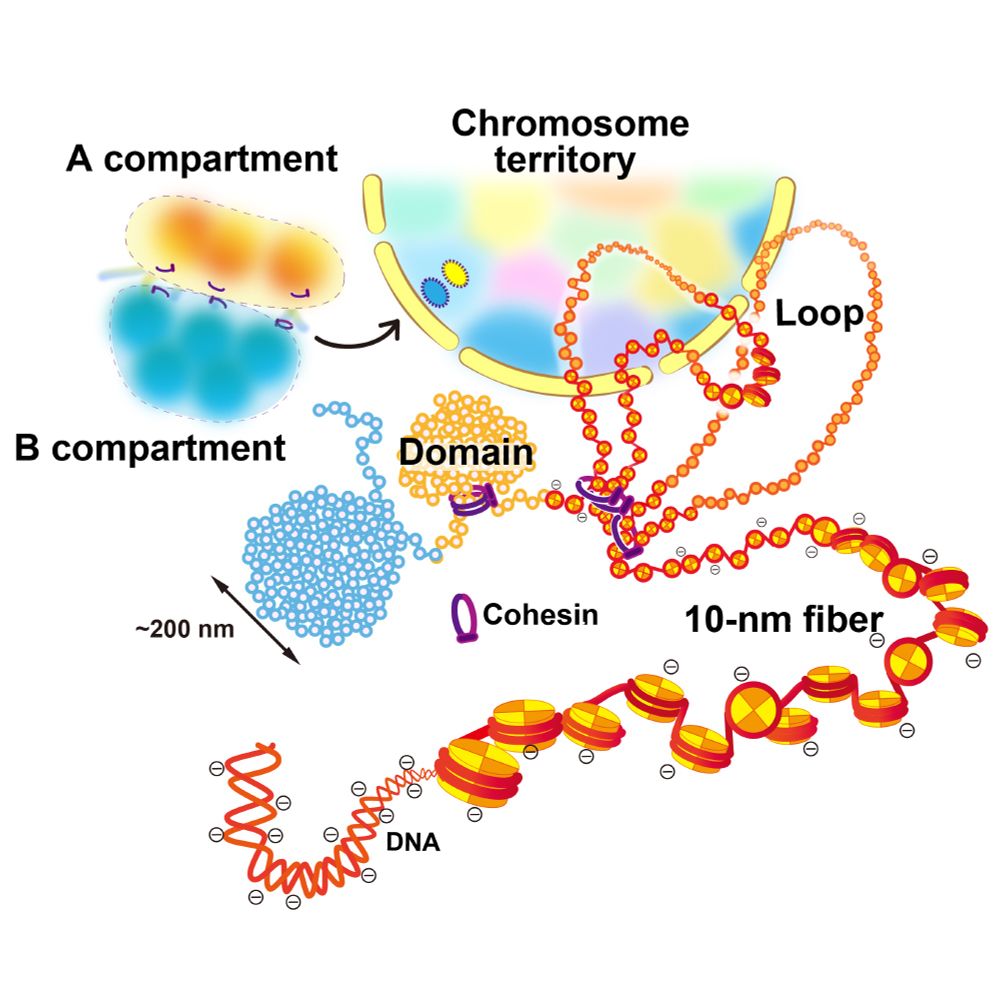
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
www.nature.com/articles/s41...
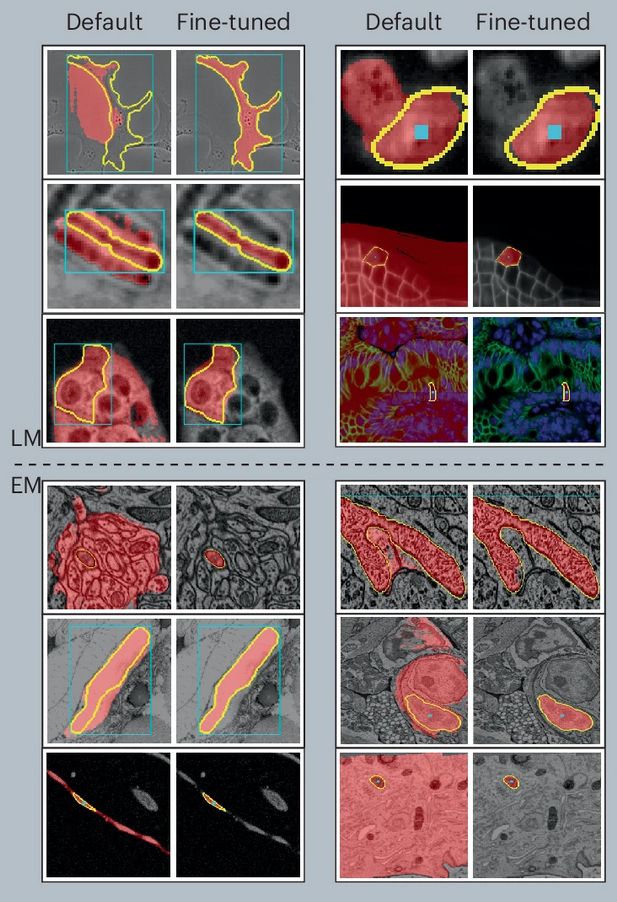
www.nature.com/articles/s41...
And thanks ChatGPT for haikuing the abstract
And thanks ChatGPT for haikuing the abstract
www.biorxiv.org/content/10.1...
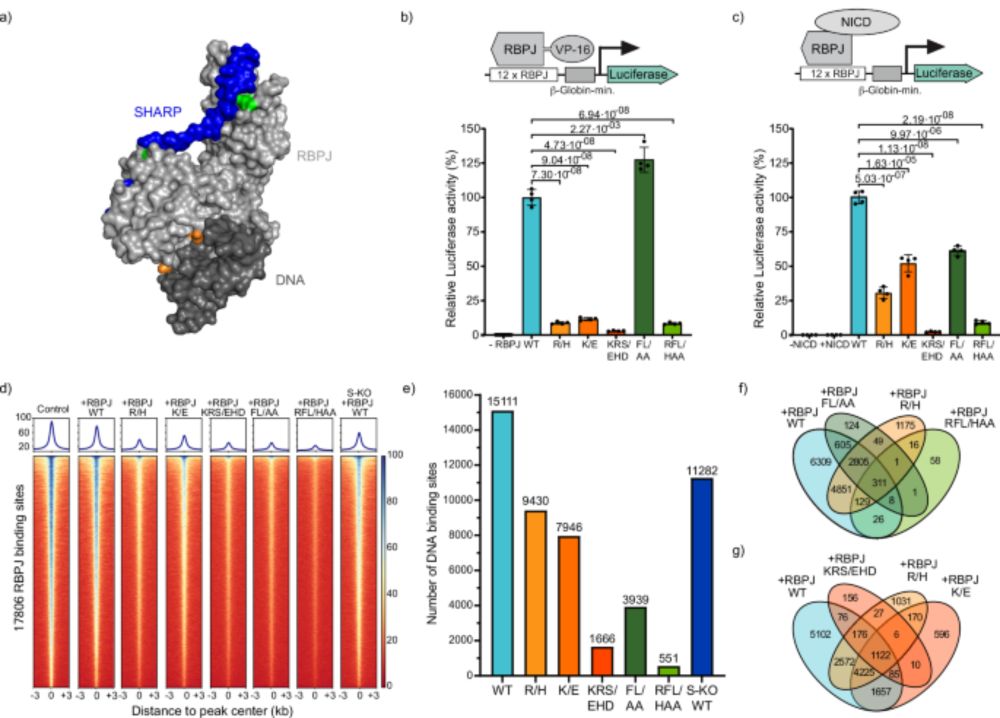
▶️40-fold flanking sequence effect on TF binding
▶️Motif recognition beats kinetics
▶️Minutes-long residence times
▶️Models of TF-nucleosome competiton




▶️40-fold flanking sequence effect on TF binding
▶️Motif recognition beats kinetics
▶️Minutes-long residence times
▶️Models of TF-nucleosome competiton

