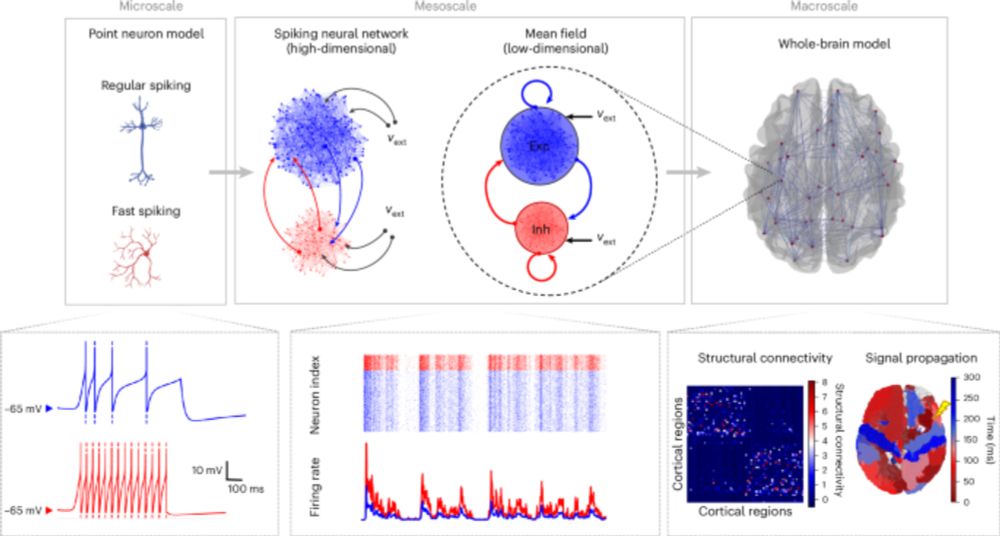
The challenge:
📈 create an algorithm that matches fluorescent images of genetic tools to the most likely spatial transcriptomic cell types
💻 present it as part of a user-friendly tool

The challenge:
📈 create an algorithm that matches fluorescent images of genetic tools to the most likely spatial transcriptomic cell types
💻 present it as part of a user-friendly tool
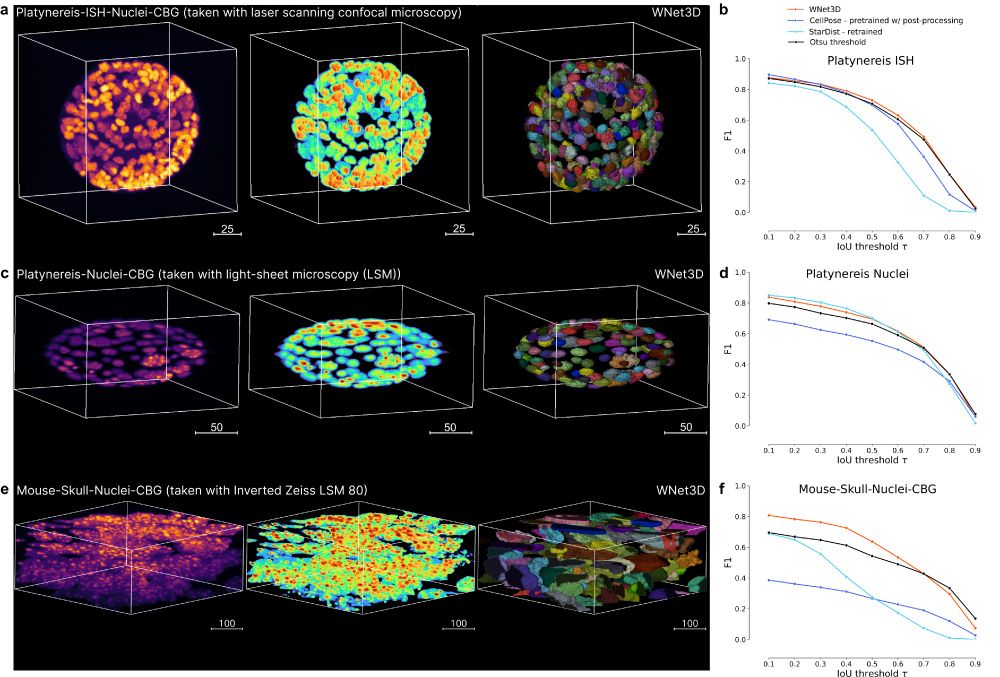
👩💻 pip install napari-cellseg3d==0.2.2
📚 biorxiv.org/content/10.1...
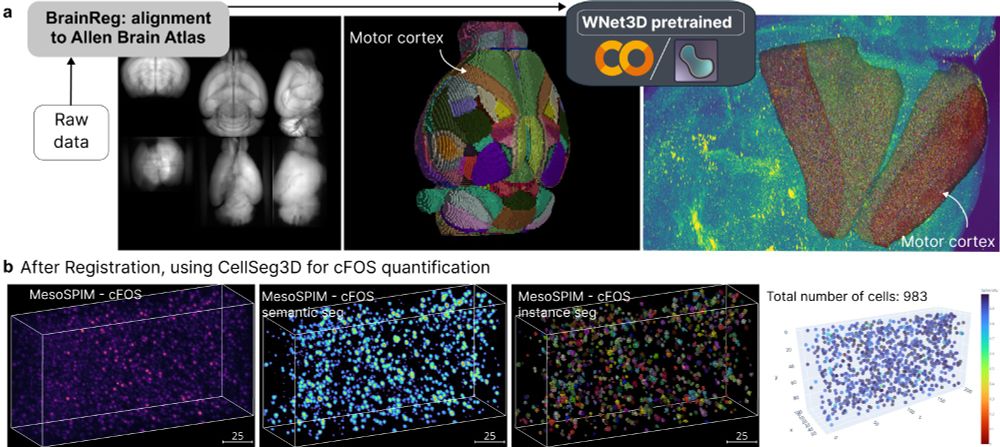
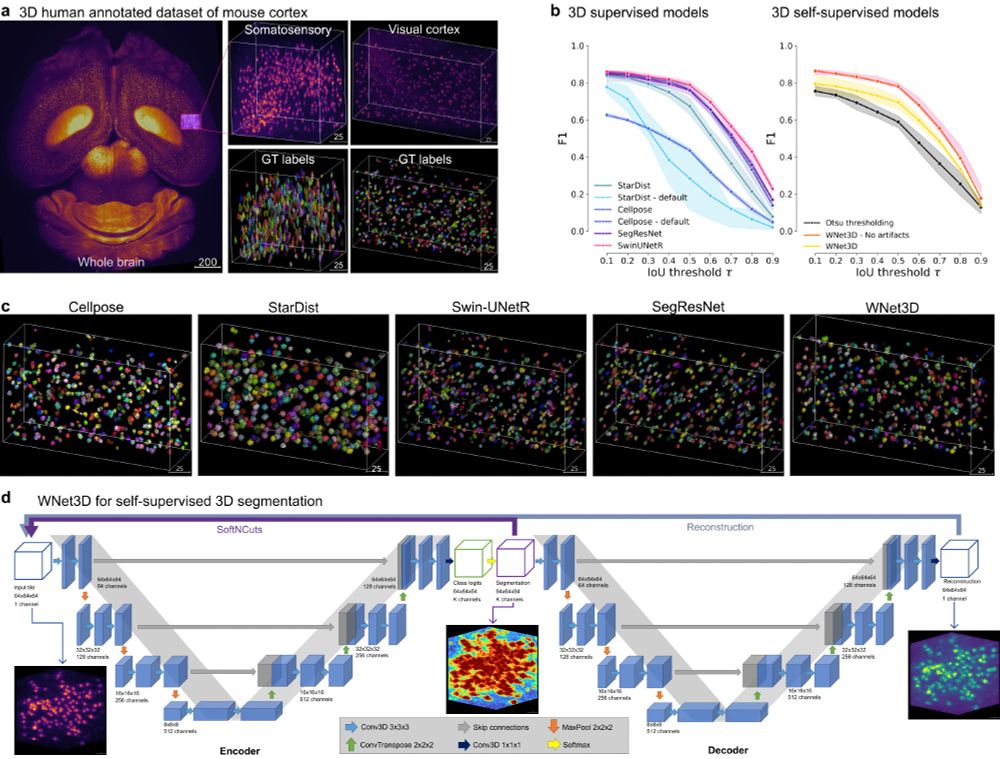
We extended benchmarking, new videos, extended text, and a demo on cFOS 👋
Join us for a webinar on how to use it in your #neuroscience research, incl. studies using our Alzheimer's datasets.
📆 Dec. 18, 9:30am PT
💻 Webinar
🎟️ alleninstitute.org/events/cell_...
#neuroskyence

Join us for a webinar on how to use it in your #neuroscience research, incl. studies using our Alzheimer's datasets.
📆 Dec. 18, 9:30am PT
💻 Webinar
🎟️ alleninstitute.org/events/cell_...
#neuroskyence
By none other than one of my favorite theoretical biologists. Jumps all the way to the top of the reading list!
And free access for 2 weeks (??), so get it right away.
#biology
www.cambridge.org/core/element...

By none other than one of my favorite theoretical biologists. Jumps all the way to the top of the reading list!
And free access for 2 weeks (??), so get it right away.
#biology
www.cambridge.org/core/element...
graph-tool.skewed.de
Graph-tool is a comprehensive and efficient Python library to work with networks, including structural, dynamical, and statistical algorithms, as well as visualization. 1/N
#networkscience
graph-tool.skewed.de
Graph-tool is a comprehensive and efficient Python library to work with networks, including structural, dynamical, and statistical algorithms, as well as visualization. 1/N
#networkscience
Can't wait? Sign up for email updates below!
info.addgene.org/pac...

www.biorxiv.org/content/10.1...
https://buff.ly/3VbDo1X

https://buff.ly/3VbDo1X






