us-hupo.org/Distinguishe...

us-hupo.org/Distinguishe...
us-hupo.org/Computationa...

us-hupo.org/Computationa...
Also looking at #Disease modelling. Fantastic #TourDForce Valuable bench marking and insight
@dschweppe.bsky.social #SCBEMs


Also looking at #Disease modelling. Fantastic #TourDForce Valuable bench marking and insight
@dschweppe.bsky.social #SCBEMs
www.nature.com/articles/s41...

www.nature.com/articles/s41...

The new pre-print from @riley-research.bsky.social seeks to answer this question using the Proteograph technology from Seer Inc.
Check out this lovely study on glycoproteins NP-enriched from biofluids here: chemrxiv.org/engage/chemr...

The new pre-print from @riley-research.bsky.social seeks to answer this question using the Proteograph technology from Seer Inc.
Check out this lovely study on glycoproteins NP-enriched from biofluids here: chemrxiv.org/engage/chemr...
Nova - lightweight dev library
schweppelab.github.io/Nova/ pubs.acs.org/doi/full/10....
Helios - a unified instrument API now @ JPR
github.com/SchweppeLab/...

Nova - lightweight dev library
schweppelab.github.io/Nova/ pubs.acs.org/doi/full/10....
Helios - a unified instrument API now @ JPR
github.com/SchweppeLab/...


---
#proteomics #prot-other

---
#proteomics #prot-other
With the complexity of glycoproteomics and how little we still know, Tim Veth puts out a rallying cry to speed up development in this promising field.
Learn more about Tim & read our discussion: bit.ly/3IXpo8B

With the complexity of glycoproteomics and how little we still know, Tim Veth puts out a rallying cry to speed up development in this promising field.
Learn more about Tim & read our discussion: bit.ly/3IXpo8B
---
#proteomics #prot-paper

---
#proteomics #prot-paper

www.nature.com/articles/s41...
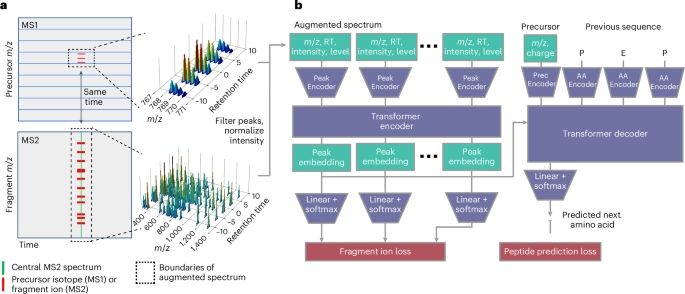
www.nature.com/articles/s41...
Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!



Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!



Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!
pubs.acs.org/doi/full/10....
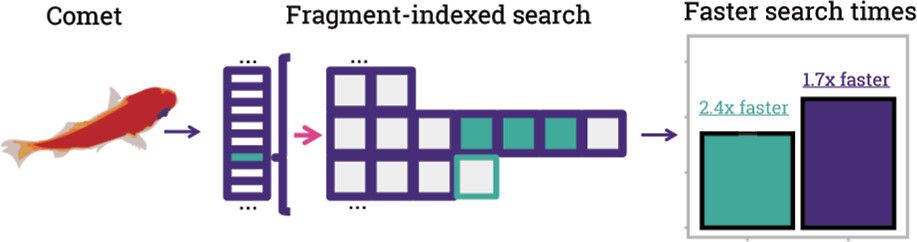
pubs.acs.org/doi/full/10....
www.nature.com/articles/s41...
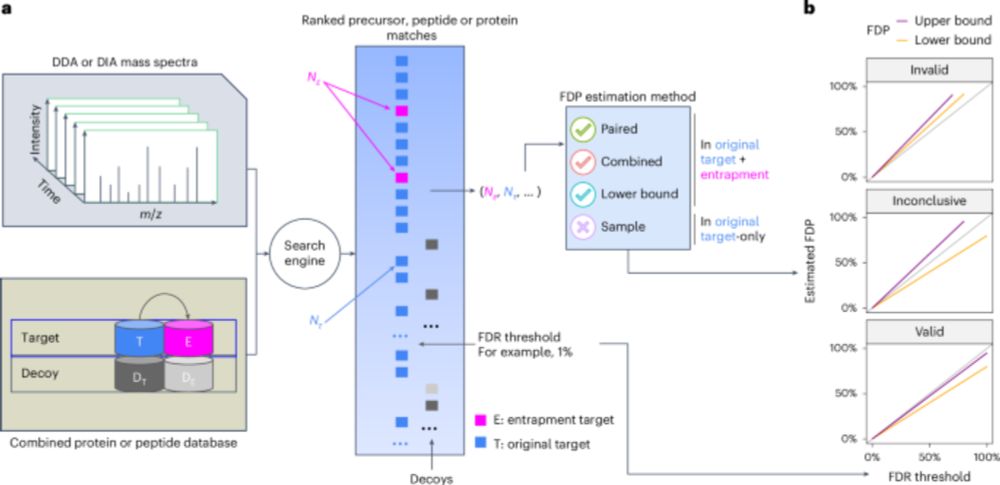
www.nature.com/articles/s41...
Photo credit to Katarina for capturing the moment!

Photo credit to Katarina for capturing the moment!


consider the Real time Mass Spectrometry Workshop featuring speakers and panelists Sarah Sipe, Aarthie Senathirajah, and Manuel Peris Diaz.

consider the Real time Mass Spectrometry Workshop featuring speakers and panelists Sarah Sipe, Aarthie Senathirajah, and Manuel Peris Diaz.
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...



