
Manuel Lessi: this work “combines the strengths of primary cultures and pooled CRISPRi perturbation”. #preprint Jingwen Ding & team @brainevodevo.bsky.social
#preLight ⬇️
prelights.biologists.com/highlights/d...

Manuel Lessi: this work “combines the strengths of primary cultures and pooled CRISPRi perturbation”. #preprint Jingwen Ding & team @brainevodevo.bsky.social
#preLight ⬇️
prelights.biologists.com/highlights/d...


We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
We compared deer mice evolved in forest vs prairie habitats. We found that forest mice have:
(1) more corticospinal neurons (CSNs)
(2) better hand dexterity
(3) more dexterous climbing, which is linked to CSN number🧵
Work by @Jeff Moore now at USC, a collaboration with @Sam Pfaff lab


Work by @Jeff Moore now at USC, a collaboration with @Sam Pfaff lab
New work on HAQERs, by
@rimangan.bsky.social Yanting Luo Craig Lowe @debbysilver.bsky.social @manoliskellis.bsky.social & colleagues
🧪🧬
www.biorxiv.org/content/10.1...

New work on HAQERs, by
@rimangan.bsky.social Yanting Luo Craig Lowe @debbysilver.bsky.social @manoliskellis.bsky.social & colleagues
🧪🧬
www.biorxiv.org/content/10.1...
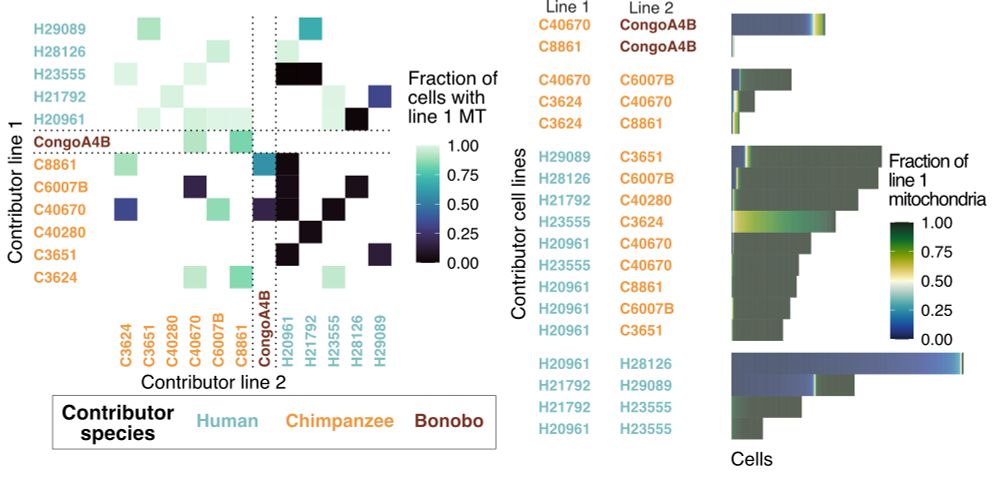
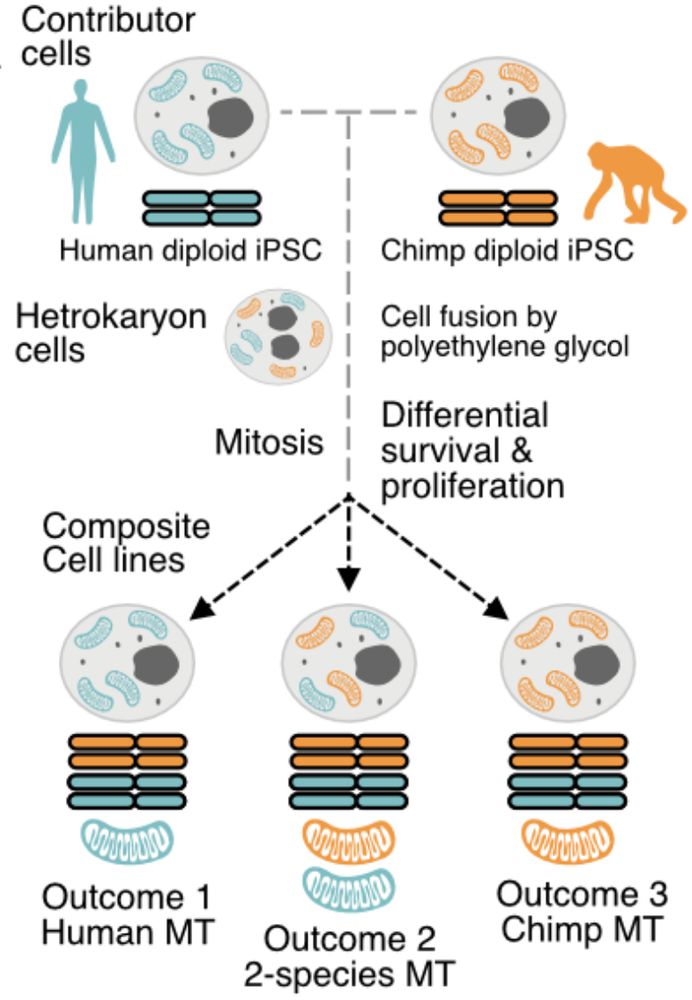
He’ll present Cellbouncer, a new bioinformatic tool for pooled single-cell processing that yields insights into hominid evolution 🧬
More about the lab 👉 www.pollenlab.org

He’ll present Cellbouncer, a new bioinformatic tool for pooled single-cell processing that yields insights into hominid evolution 🧬
More about the lab 👉 www.pollenlab.org
Pure vandalism.
This could never happen in a country with functioning checks and balances.

Pure vandalism.
This could never happen in a country with functioning checks and balances.
@aydoganlab.bsky.social @jamesbriscoe.bsky.social @brunetlab.bsky.social @ebisuyamiki.bsky.social @mwdorr.bsky.social @beyerlab.bsky.social @franckp.bsky.social @janereznick.bsky.social @olmedo-lab.bsky.social @perez-carrasco.bsky.social
meetings.embo.org/event/25-dev...


www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

