Alex Pollen
@brainevodevo.bsky.social
Studying specializations and vulnerabilities of human brain development
We begin by performing cross-species analysis of developing initial classes of both striatal and cortical inhibitory neuron populations by integrating single cell sequencing data from 8 mammals, spanning from primates to marsupials.

November 7, 2025 at 6:06 PM
We begin by performing cross-species analysis of developing initial classes of both striatal and cortical inhibitory neuron populations by integrating single cell sequencing data from 8 mammals, spanning from primates to marsupials.
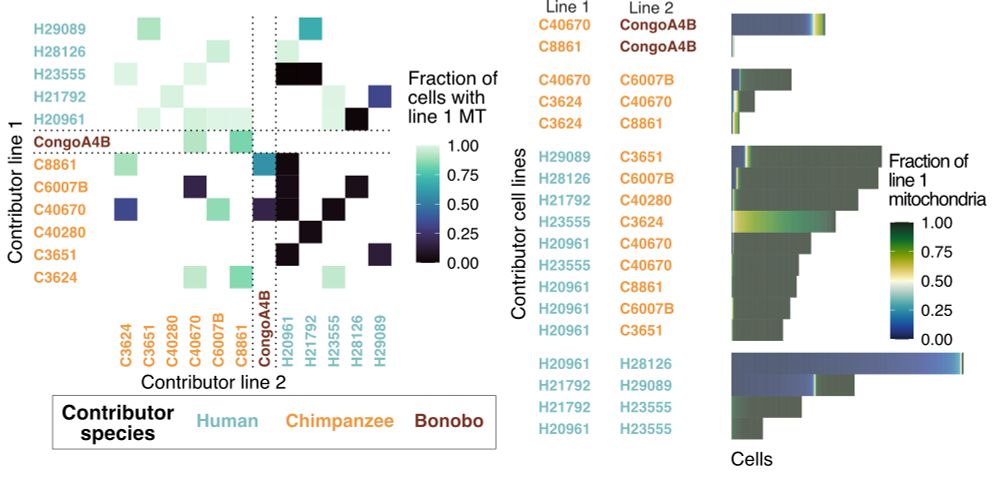
Our analysis revealed that human and bonobo mitochondria typically outcompete those from chimpanzee, but we also identified a fraction of cells where chimpanzee mitochondria win and cells where both mitochondria survive.
March 24, 2025 at 10:09 PM
Our analysis revealed that human and bonobo mitochondria typically outcompete those from chimpanzee, but we also identified a fraction of cells where chimpanzee mitochondria win and cells where both mitochondria survive.
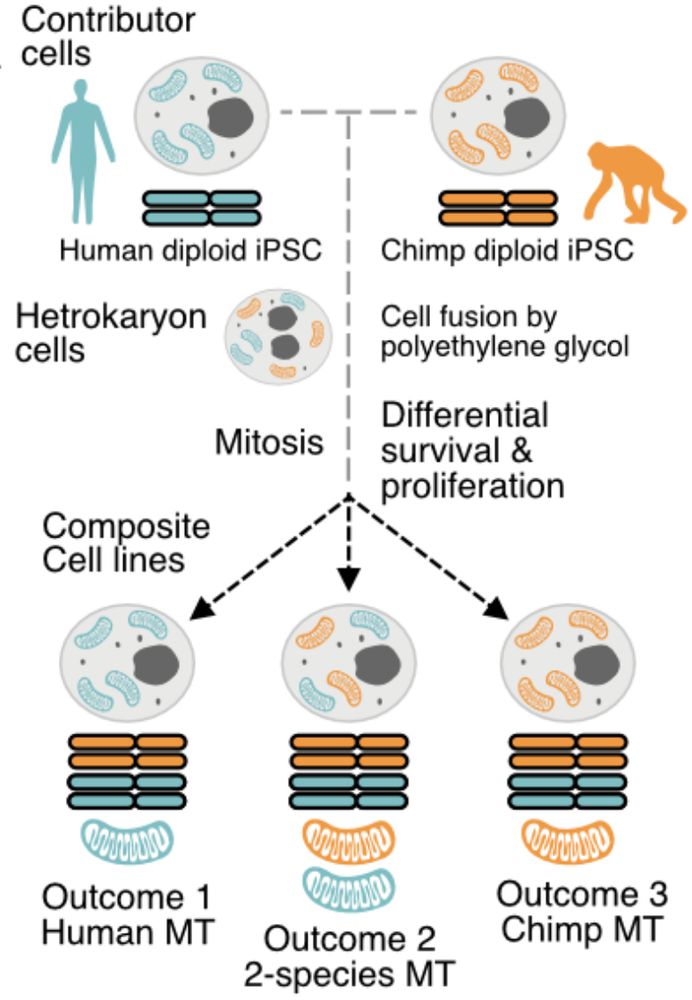
We put CellBouncer to the test with a challenging demultiplexing problem of assigning 24 hominid tetraploid composite cell lines generated by Bryan Pavlovic to both individuals-of-origin and identifying the mitochondrial haplotypes present.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
March 24, 2025 at 10:07 PM
We put CellBouncer to the test with a challenging demultiplexing problem of assigning 24 hominid tetraploid composite cell lines generated by Bryan Pavlovic to both individuals-of-origin and identifying the mitochondrial haplotypes present.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
CellBouncer tools were crucial for our study of dopaminergic evolution that included ventral midbrain organoids with up to 17 individuals from 4 species, led by Sara Nolbrant, @jenellewallace.bsky.social, and Jingwen Ding:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
March 24, 2025 at 10:04 PM
CellBouncer tools were crucial for our study of dopaminergic evolution that included ventral midbrain organoids with up to 17 individuals from 4 species, led by Sara Nolbrant, @jenellewallace.bsky.social, and Jingwen Ding:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Finally, CellBouncer harmonizes independent metrics for doublet identification to produce a global doublet rate

March 24, 2025 at 10:02 PM
Finally, CellBouncer harmonizes independent metrics for doublet identification to produce a global doublet rate
CellBouncer assigns sgRNAs and other perturbation tags to cell-of-origin, accounting for low coverage and increased tag background
March 24, 2025 at 10:01 PM
CellBouncer assigns sgRNAs and other perturbation tags to cell-of-origin, accounting for low coverage and increased tag background
CellBouncer also provides independent validation methods for statistical confidence in assignments, including a tool for determining individual proportion in bulk (and pseudobulk) RNA-seq data
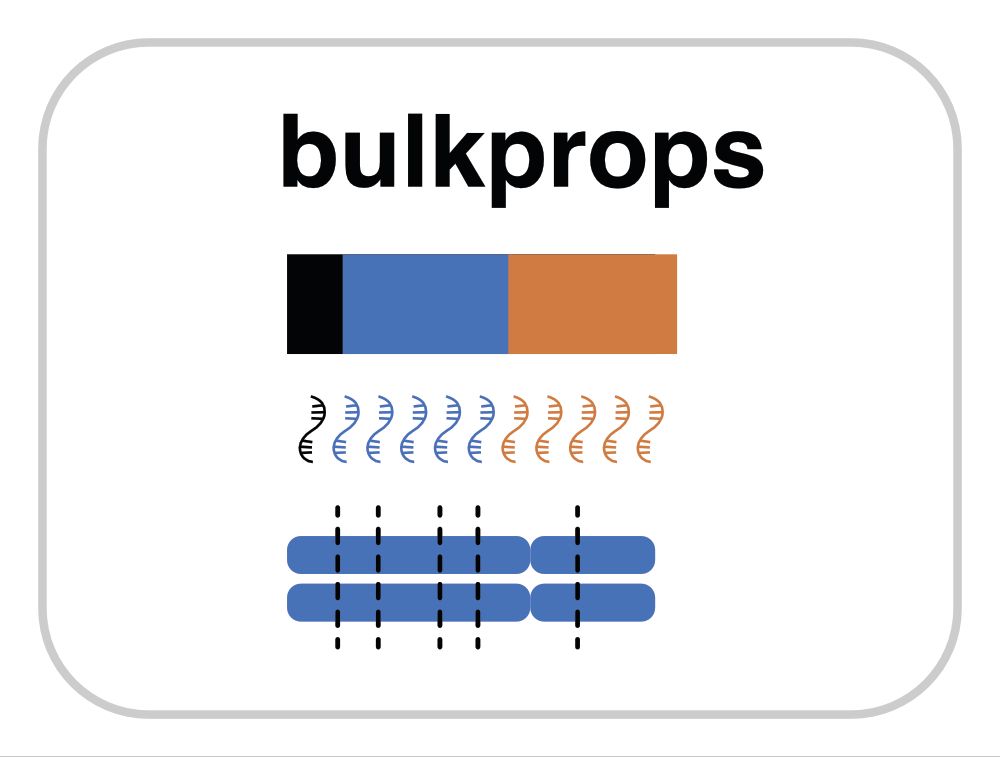
March 24, 2025 at 9:59 PM
CellBouncer also provides independent validation methods for statistical confidence in assignments, including a tool for determining individual proportion in bulk (and pseudobulk) RNA-seq data
Importantly, CellBouncer uses sequence variation as an external ground-truth for ambient RNA determination, improving individual-of-origin assignments and enabling cell level corrections, while providing insights into the nature and sources of ambient RNA.
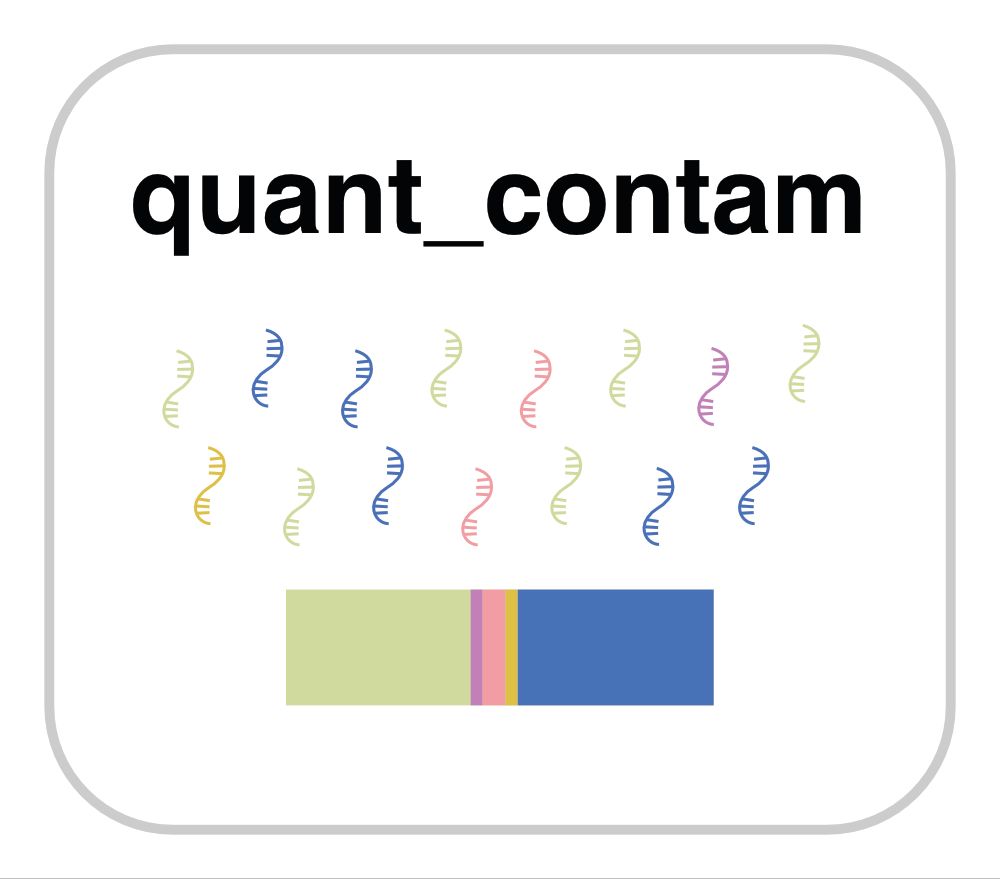
March 24, 2025 at 9:57 PM
Importantly, CellBouncer uses sequence variation as an external ground-truth for ambient RNA determination, improving individual-of-origin assignments and enabling cell level corrections, while providing insights into the nature and sources of ambient RNA.
Genotype-free assignments for an unknown number of individuals by discovery of mitochondrial haplotypes
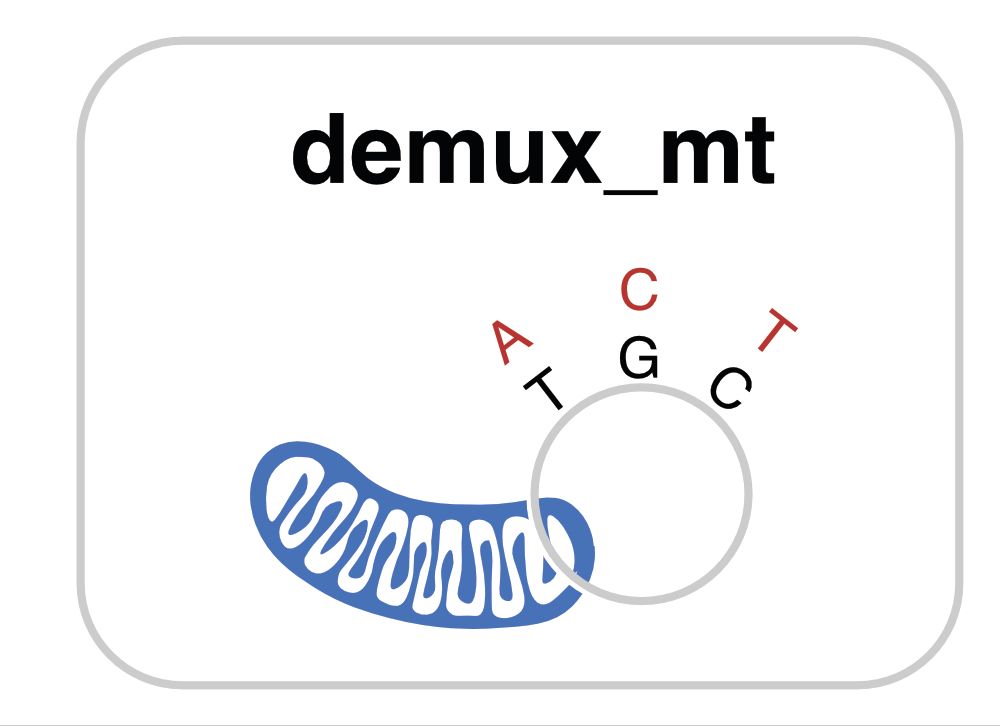
March 24, 2025 at 9:56 PM
Genotype-free assignments for an unknown number of individuals by discovery of mitochondrial haplotypes
Individual-of-origin assignments for cells that account for deep population structure, scale across large SNP sets, and are robust to ambient RNA
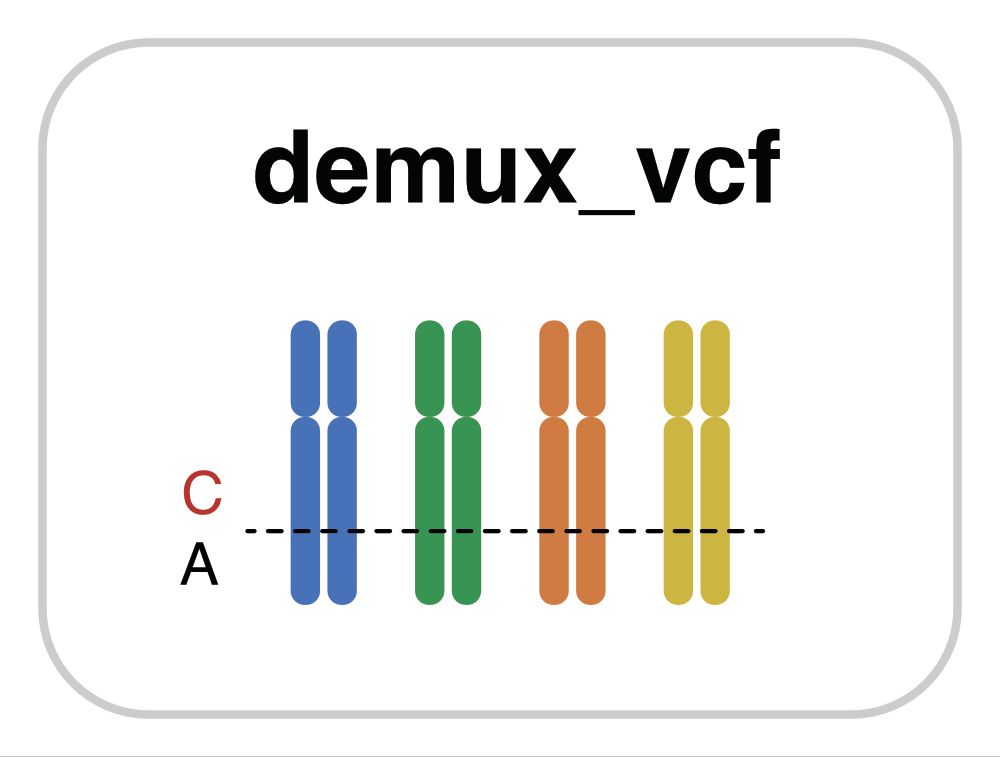
March 24, 2025 at 9:56 PM
Individual-of-origin assignments for cells that account for deep population structure, scale across large SNP sets, and are robust to ambient RNA
Alignment-free rapid species assignments with an underlying statistical model
March 24, 2025 at 9:55 PM
Alignment-free rapid species assignments with an underlying statistical model
Introducing CellBouncer, a unified demultiplexing toolkit built by nkschaefer.bsky.social to check IDs and keep the riff raff out of single cell genomics datasets, including by using genetic variation as an external ground-truth for ambient RNA removal:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

March 24, 2025 at 9:48 PM
Introducing CellBouncer, a unified demultiplexing toolkit built by nkschaefer.bsky.social to check IDs and keep the riff raff out of single cell genomics datasets, including by using genetic variation as an external ground-truth for ambient RNA removal:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
@jenellewallace.bsky.social
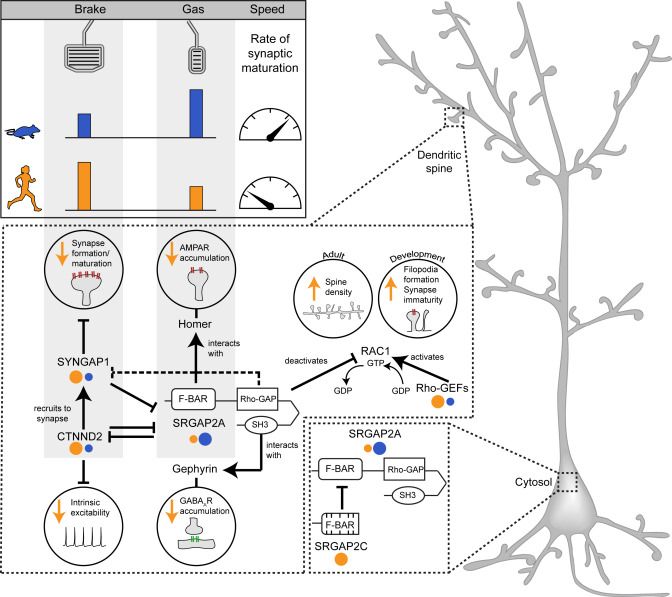
led a preview of two outstanding papers from the labs of Cecile Charrier, Pierre Vanderhaeghen, and @franckp.bsky.social that connect regulation of the tempo of synaptogenesis by the human-specific gene SRGAP2C to ASD-linked genes: authors.elsevier.com/a/1k3583BtfH...
led a preview of two outstanding papers from the labs of Cecile Charrier, Pierre Vanderhaeghen, and @franckp.bsky.social that connect regulation of the tempo of synaptogenesis by the human-specific gene SRGAP2C to ASD-linked genes: authors.elsevier.com/a/1k3583BtfH...

November 12, 2024 at 10:35 PM
@jenellewallace.bsky.social
led a preview of two outstanding papers from the labs of Cecile Charrier, Pierre Vanderhaeghen, and @franckp.bsky.social that connect regulation of the tempo of synaptogenesis by the human-specific gene SRGAP2C to ASD-linked genes: authors.elsevier.com/a/1k3583BtfH...
led a preview of two outstanding papers from the labs of Cecile Charrier, Pierre Vanderhaeghen, and @franckp.bsky.social that connect regulation of the tempo of synaptogenesis by the human-specific gene SRGAP2C to ASD-linked genes: authors.elsevier.com/a/1k3583BtfH...
Building on our recent review, Tyler Fair performed CRISPRi-based genetic screens to assign human-specific deletions to molecular and cellular phenotypes:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

December 29, 2023 at 6:49 PM
Building on our recent review, Tyler Fair performed CRISPRi-based genetic screens to assign human-specific deletions to molecular and cellular phenotypes:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

