📍University of Cambridge @cambridge-uni.bsky.social | @UKDRI | 🔬developing live cell biosensing tech
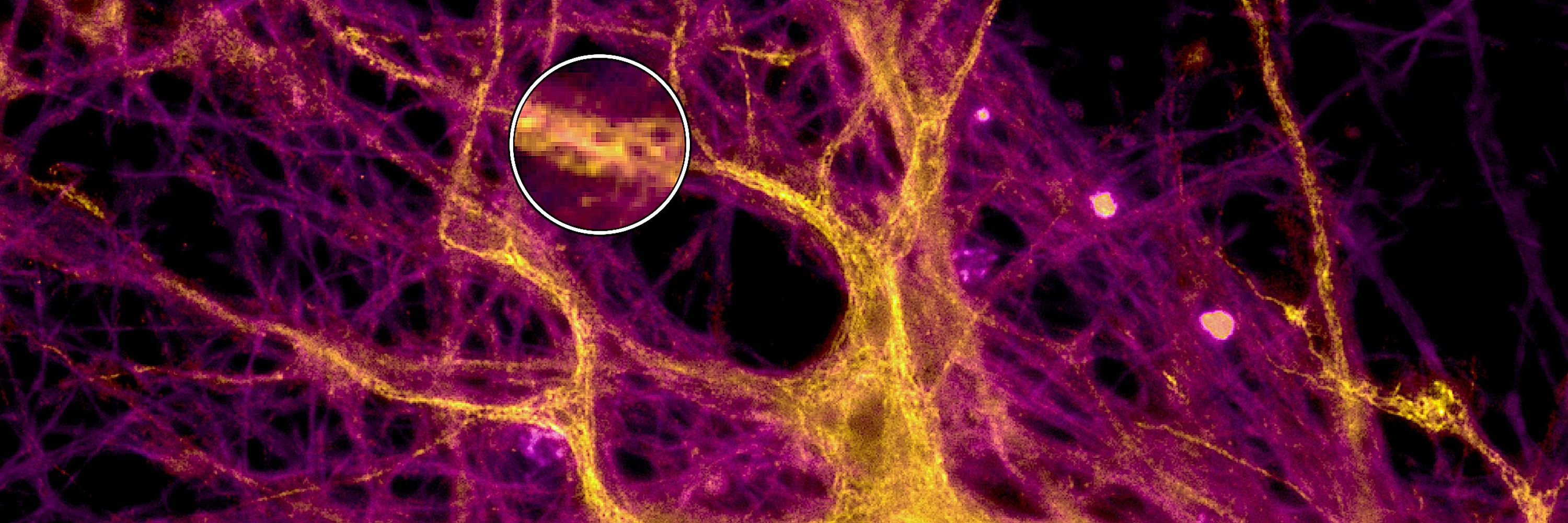
With it we resolve with sub-organelle res. e.g. BACE1 amyloidogenic APP cleavage #Alzheimers, ER exit events, map nanobody binding in realtime in ER/organelles 🔬🧠#SingleMolecule
rdcu.be/e3Ris
Ribosomal RNA expansion segments mediate the oligomerization of inactive animal ribosomes | Science www.science.org/doi/10.1126/...

Ribosomal RNA expansion segments mediate the oligomerization of inactive animal ribosomes | Science www.science.org/doi/10.1126/...
🙏
🙏
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
🙏
🙏
www.nature.com/articles/s41...

www.nature.com/articles/s41...
With it we resolve with sub-organelle res. e.g. BACE1 amyloidogenic APP cleavage #Alzheimers, ER exit events, map nanobody binding in realtime in ER/organelles 🔬🧠#SingleMolecule
rdcu.be/e3Ris
With it we resolve with sub-organelle res. e.g. BACE1 amyloidogenic APP cleavage #Alzheimers, ER exit events, map nanobody binding in realtime in ER/organelles 🔬🧠#SingleMolecule
rdcu.be/e3Ris
bit.ly/40dNHEE

bit.ly/40dNHEE
With it we resolve with sub-organelle res. e.g. BACE1 amyloidogenic APP cleavage #Alzheimers, ER exit events, map nanobody binding in realtime in ER/organelles 🔬🧠#SingleMolecule
rdcu.be/e3Ris
With it we resolve with sub-organelle res. e.g. BACE1 amyloidogenic APP cleavage #Alzheimers, ER exit events, map nanobody binding in realtime in ER/organelles 🔬🧠#SingleMolecule
rdcu.be/e3Ris
Highlight: CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis www.cell.com/cell/fulltex...
Highlight: CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis www.cell.com/cell/fulltex...
DNA-protein cross-links promote cGAS-STING–driven premature aging and embryonic lethality | Science www.science.org/doi/10.1126/...

DNA-protein cross-links promote cGAS-STING–driven premature aging and embryonic lethality | Science www.science.org/doi/10.1126/...
TuroID attached to inactive Cas to identify transcription regulators 👇
www.nature.com/articles/s41...

TuroID attached to inactive Cas to identify transcription regulators 👇
www.nature.com/articles/s41...
Abstract submission/Registration by 31 May/15 Jun
https://meetings.embo.org/event/26-axonal-degeneration
#EMBOAxonalRegeneration #EMBOevents #meeting 🧪

Abstract submission/Registration by 31 May/15 Jun
https://meetings.embo.org/event/26-axonal-degeneration
#EMBOAxonalRegeneration #EMBOevents #meeting 🧪
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....









nature.com/articles/s41...
rdcu.be/eOfBH
This new Review by @dannomura.bsky.social et al. discusses the rapidly expanding landscape of therapeutic approaches based on inducing proximity between proteins, including targeted protein degraders and more

nature.com/articles/s41...
rdcu.be/eOfBH
This new Review by @dannomura.bsky.social et al. discusses the rapidly expanding landscape of therapeutic approaches based on inducing proximity between proteins, including targeted protein degraders and more
www.biorxiv.org/content/10.1...
#Neurodegeneration #NeuroCellBiology #Calcium #bioRxiv
www.biorxiv.org/content/10.1...
#Neurodegeneration #NeuroCellBiology #Calcium #bioRxiv


