
Folks @ Retro achieved 50x improvement on iPSC generation using engineered versions of OSKM over wild-type!
🧪🧬💻👾
openai.com/index/accele...

Folks @ Retro achieved 50x improvement on iPSC generation using engineered versions of OSKM over wild-type!
🧪🧬💻👾
openai.com/index/accele...
By learning the latent representation of phenotype space, they speed up and directly learn the phenotype transition --> perturbagen mapping.
Super cool!!
www.nature.com/articles/s41...
#AI #drugdiscovery #cancer

By learning the latent representation of phenotype space, they speed up and directly learn the phenotype transition --> perturbagen mapping.
Super cool!!
www.nature.com/articles/s41...
#AI #drugdiscovery #cancer
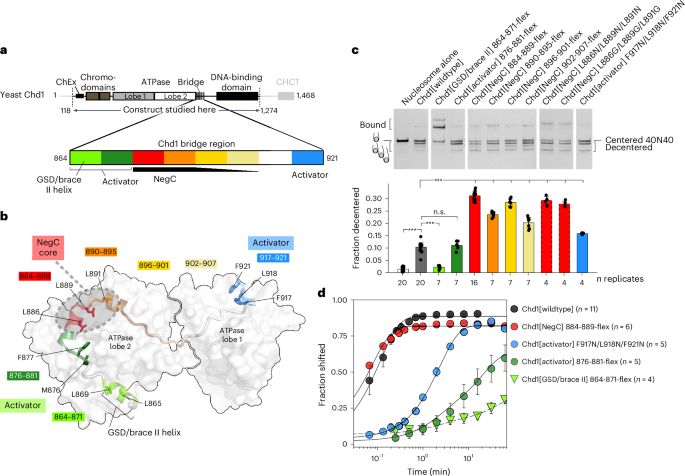

▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
#HarvardImmunology 🧪

#HarvardImmunology 🧪
Many of the DFCI Seminars in Oncology are recorded and open to the public, enjoy!
seminarsinoncology.dana-farber.org/2025.html
🧪
Many of the DFCI Seminars in Oncology are recorded and open to the public, enjoy!
seminarsinoncology.dana-farber.org/2025.html
🧪
This article makes huge, speculative leaps about treatment for a disease based only on correlative findings.
Science communication matters 🧪
www.sciencealert.com/parkinsons-g...

This article makes huge, speculative leaps about treatment for a disease based only on correlative findings.
Science communication matters 🧪
I cannot recommend Dr St Laurent as a mentor highly enough, she is brilliant, passionate, kind & dedicated! 🧪
www.stlaurentlab.org/join

I cannot recommend Dr St Laurent as a mentor highly enough, she is brilliant, passionate, kind & dedicated! 🧪
www.stlaurentlab.org/join

Listen here: buff.ly/4ibPPUD

Listen here: buff.ly/4ibPPUD
File under: Things I learn in lab meetings

File under: Things I learn in lab meetings
Something like CST’s, except (1) continuously updated and (2) describing the actual modification (e.g. ‘symmetric dimethylation’, instead of ‘methylation’)…
#epigenetics #biochemisty 🧬🧪
Something like CST’s, except (1) continuously updated and (2) describing the actual modification (e.g. ‘symmetric dimethylation’, instead of ‘methylation’)…
#epigenetics #biochemisty 🧬🧪
They show asymmetric bivalency recruits repressive readers (but not H3K4me3 readers) and have emergent properties that neither of the two alone do (e.g. recruitment of KAT6B complex)!
🧪

They show asymmetric bivalency recruits repressive readers (but not H3K4me3 readers) and have emergent properties that neither of the two alone do (e.g. recruitment of KAT6B complex)!
🧪

Early detection in pancreatic cancer will be game-changer 🧪

Early detection in pancreatic cancer will be game-changer 🧪

🧪

🧪
It’s amazing the information stored in accessibility profiles! The generalized expression model they learned can accurately predict TF activity & interactions and infer cis regulatory elements
#epigenetics 🧪 🧬

It’s amazing the information stored in accessibility profiles! The generalized expression model they learned can accurately predict TF activity & interactions and infer cis regulatory elements
#epigenetics 🧪 🧬
I’m looking at the Eppendorf SciVario Twin (maybe 320?) or the Thermo G3Lab system. Any other systems I should consider?
Thanks!
🧬 🧫 🦠
I’m looking at the Eppendorf SciVario Twin (maybe 320?) or the Thermo G3Lab system. Any other systems I should consider?
Thanks!
🧬 🧫 🦠

🧬 Discover how a pioneer transcription factor-like complex infiltrates repressive heterochromatin, producing transcripts with hidden introns that kickstart RNAi-mediated heterochromatin formation www.nature.com/articles/s41...
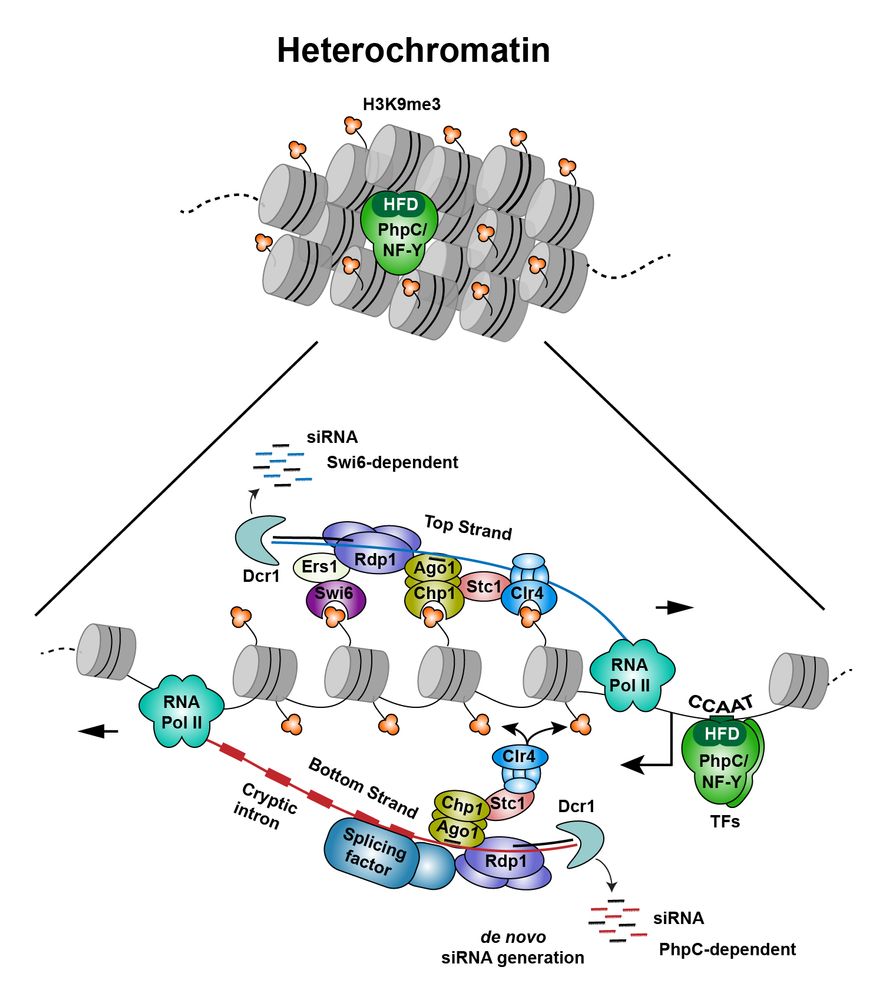
🧬 Discover how a pioneer transcription factor-like complex infiltrates repressive heterochromatin, producing transcripts with hidden introns that kickstart RNAi-mediated heterochromatin formation www.nature.com/articles/s41...
sciencedirect.com/science/arti...
A big thank you to Tom Fillot for his efforts on this and to
@hansen_lab
@marcelonollmann
for their help as editors.
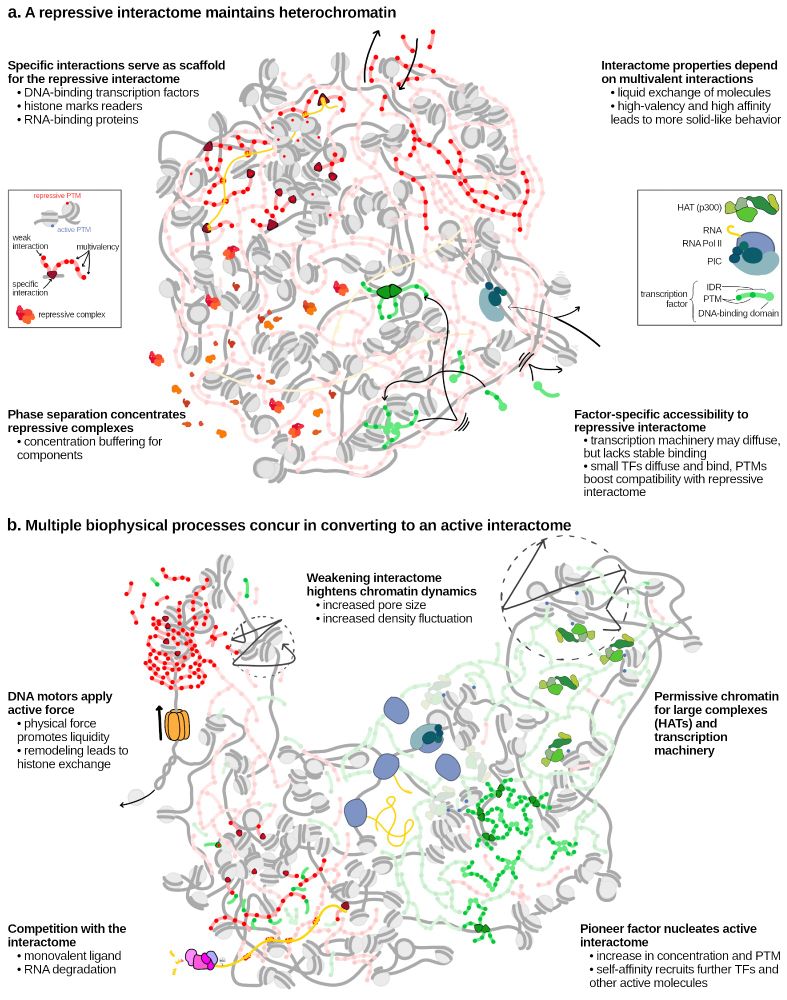
sciencedirect.com/science/arti...
A big thank you to Tom Fillot for his efforts on this and to
@hansen_lab
@marcelonollmann
for their help as editors.

