EMBL-EBI alumni, keen cyclist and father of two.
Views are my own. Reposts are not endorsement.
#RNA #ribosome #nanopore

#RNA #ribosome #nanopore
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
Join our cutting-edge research team as a molecular biologist at @nanoporetech.com HQ in Oxford.
Perfect for a fresh PhD or MSc with a couple of years’ experience. Work at the interface of chemistry, molecular/synthetic biology and AI
👉 ejnh.fa.em2.oraclecloud.com/hcmUI/Candid...

Join our cutting-edge research team as a molecular biologist at @nanoporetech.com HQ in Oxford.
Perfect for a fresh PhD or MSc with a couple of years’ experience. Work at the interface of chemistry, molecular/synthetic biology and AI
👉 ejnh.fa.em2.oraclecloud.com/hcmUI/Candid...


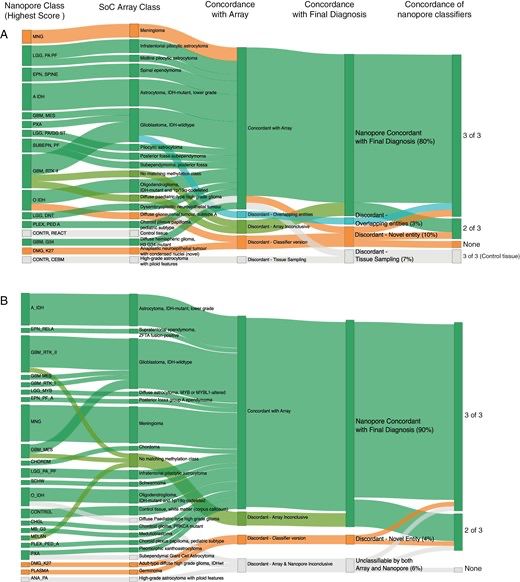
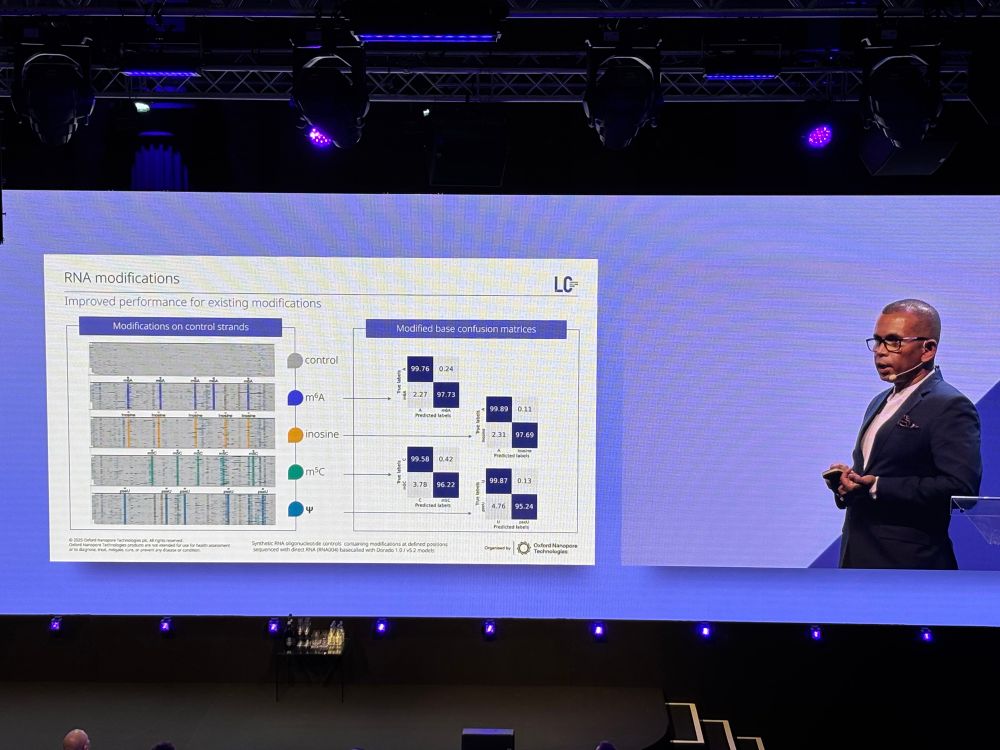
ONT is the *only* sequencing technology that detects RNA modifications directly. They're now up to 8 modifications that can be detected simultaneously.
[This is something I think ONT should be shouting from the rooftops every day]
RNA barcoding is in beta testing.
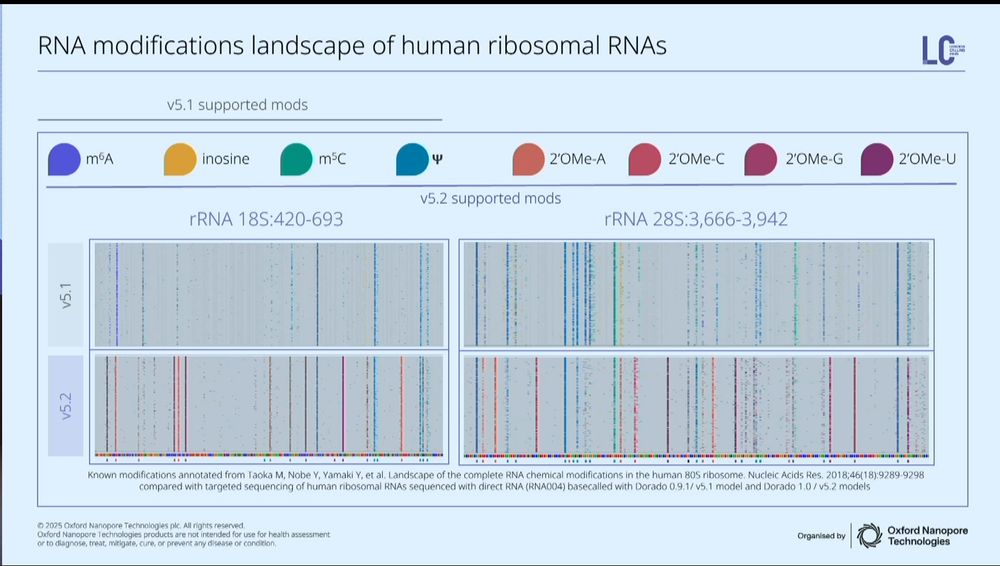
ONT is the *only* sequencing technology that detects RNA modifications directly. They're now up to 8 modifications that can be detected simultaneously.
[This is something I think ONT should be shouting from the rooftops every day]
RNA barcoding is in beta testing.




First panel-based proteomics assays, before driving towards full protein sensing in the future. #nanoporeconf

First panel-based proteomics assays, before driving towards full protein sensing in the future. #nanoporeconf

github.com/nanoporetech...


github.com/nanoporetech...
github.com/nanoporetech...



github.com/nanoporetech...

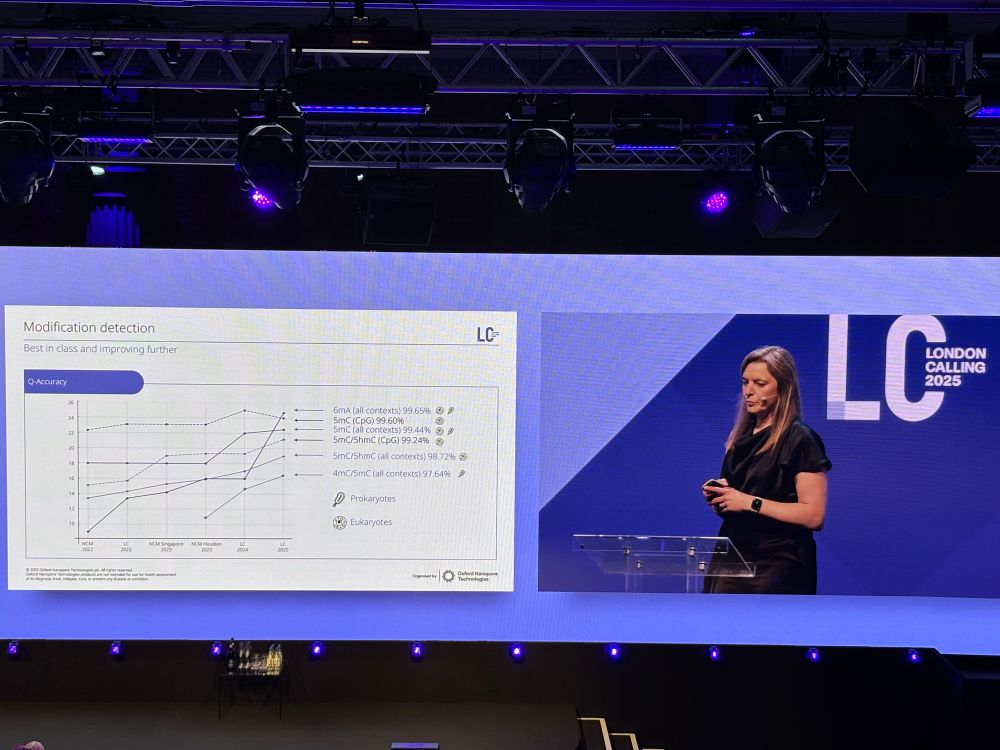
LC25 release adds v5.2 DNA/RNA models with a new suite of mod models that are higher accuracy, lower FP rates, and significantly faster.
- Introducing dorado variant 👀
- HAC error rate is down over 25%.
- New 2’Ome mod models
- Hopper & Blackwell SUP speed ups
#Nanoporeconf
LC25 release adds v5.2 DNA/RNA models with a new suite of mod models that are higher accuracy, lower FP rates, and significantly faster.
- Introducing dorado variant 👀
- HAC error rate is down over 25%.
- New 2’Ome mod models
- Hopper & Blackwell SUP speed ups
#Nanoporeconf