
Update time ! Send me a message to 1) add your lab or others 2) Correct info
docs.google.com/spreadsheets...
#Phagesky #Microsky

Update time ! Send me a message to 1) add your lab or others 2) Correct info
docs.google.com/spreadsheets...
#Phagesky #Microsky
- Carl Sagan, Cosmos

- Carl Sagan, Cosmos
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!
Today (April 29) 1 p.m. at Global Lounge in “The Commons”
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

Today (April 29) 1 p.m. at Global Lounge in “The Commons”

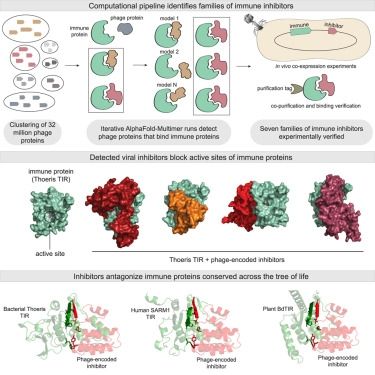
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...
Congrats Erez Yirmiya, Azita Leavitt, Gil Amitai, our collaborators at the Kranzusch lab, and coauthors
A 🧵 1/10
www.cell.com/cell/abstrac...
Learn more in a new Science study: https://scim.ag/40l2z3J

Learn more in a new Science study: https://scim.ag/40l2z3J
🌐 search.foldseek.com
🌐 search.foldseek.com
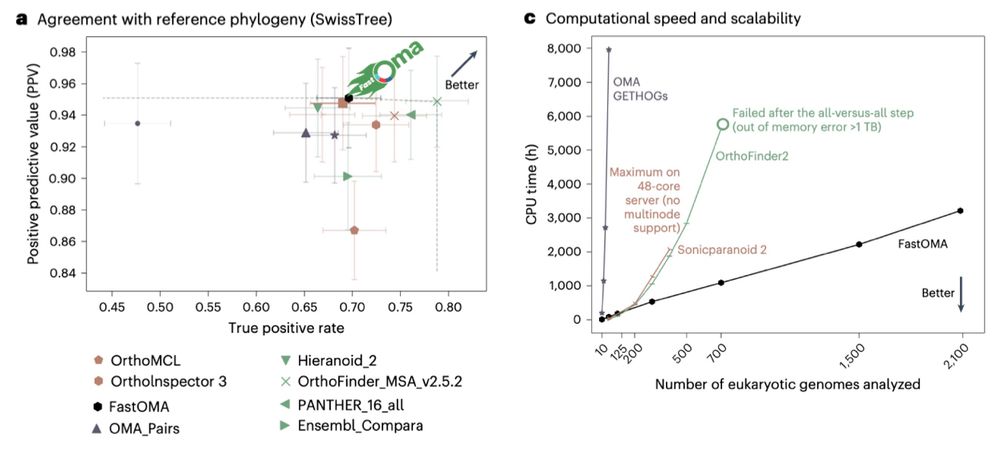
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
One with the new TREE Doctoral Landscape Awards: www.trees-dla.ac.uk/projects/int... (deadline 20th Jan)
And another with CSC: www.findaphd.com/phds/project... (deadline 29th Jan)

One with the new TREE Doctoral Landscape Awards: www.trees-dla.ac.uk/projects/int... (deadline 20th Jan)
And another with CSC: www.findaphd.com/phds/project... (deadline 29th Jan)
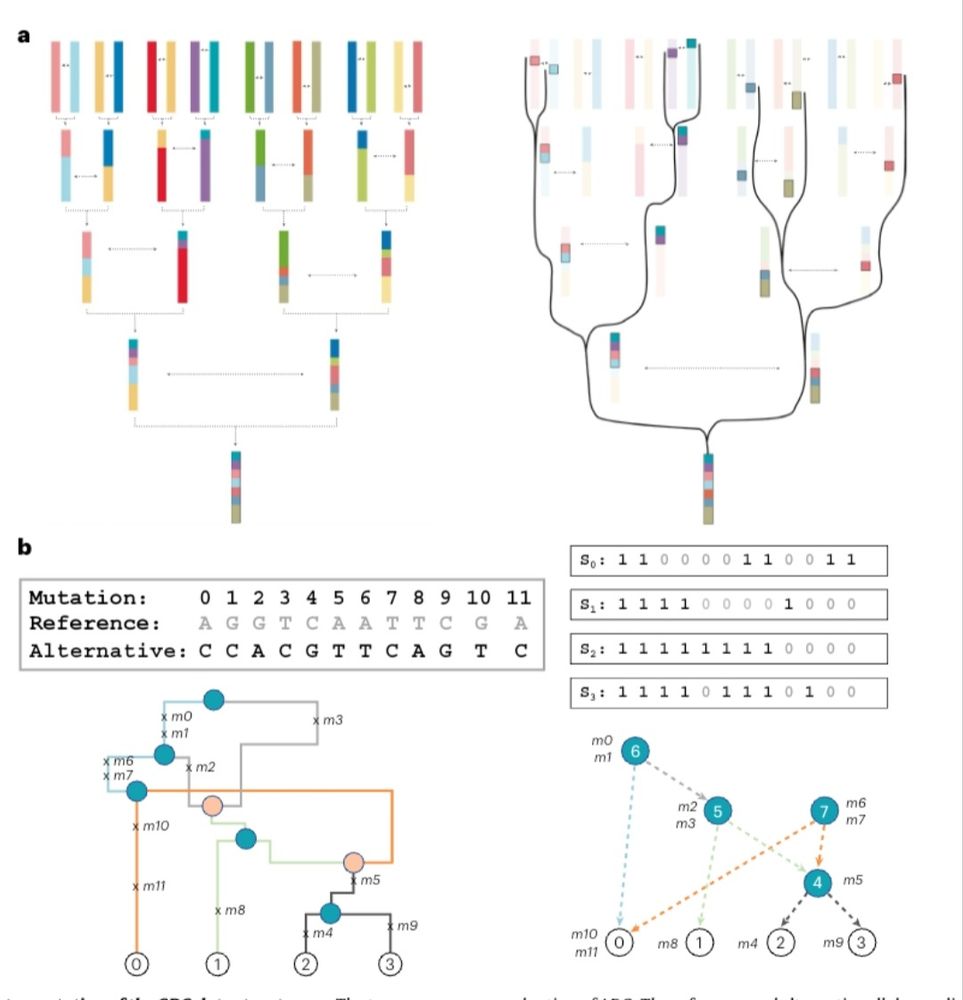
www.pnas.org/doi/full/10....

www.pnas.org/doi/full/10....


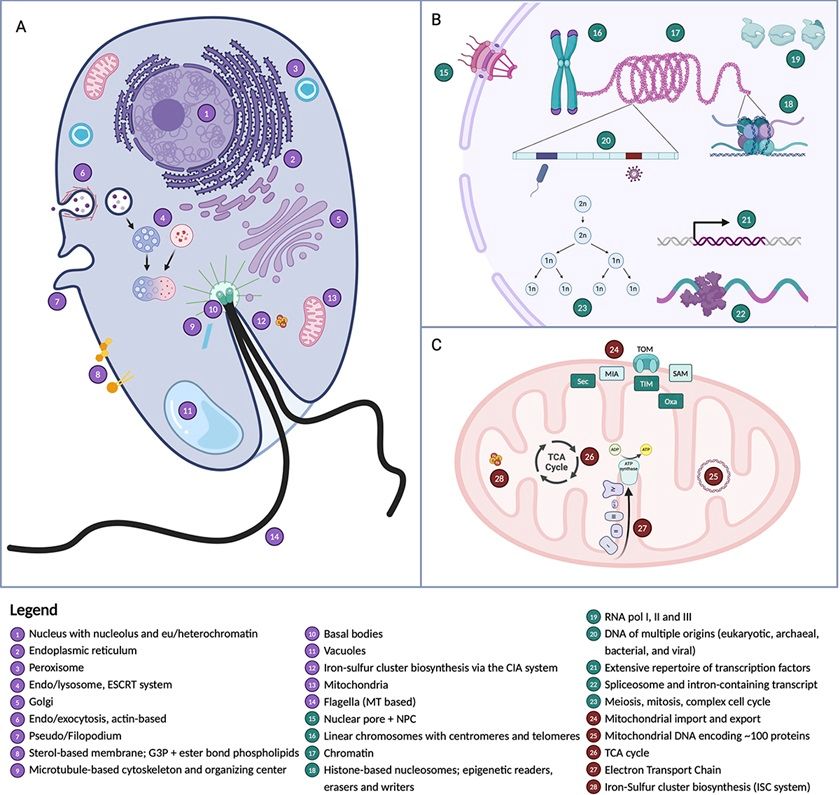
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
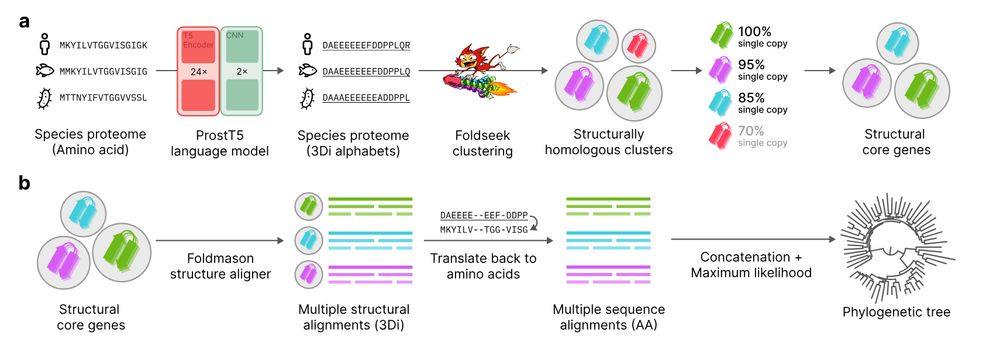
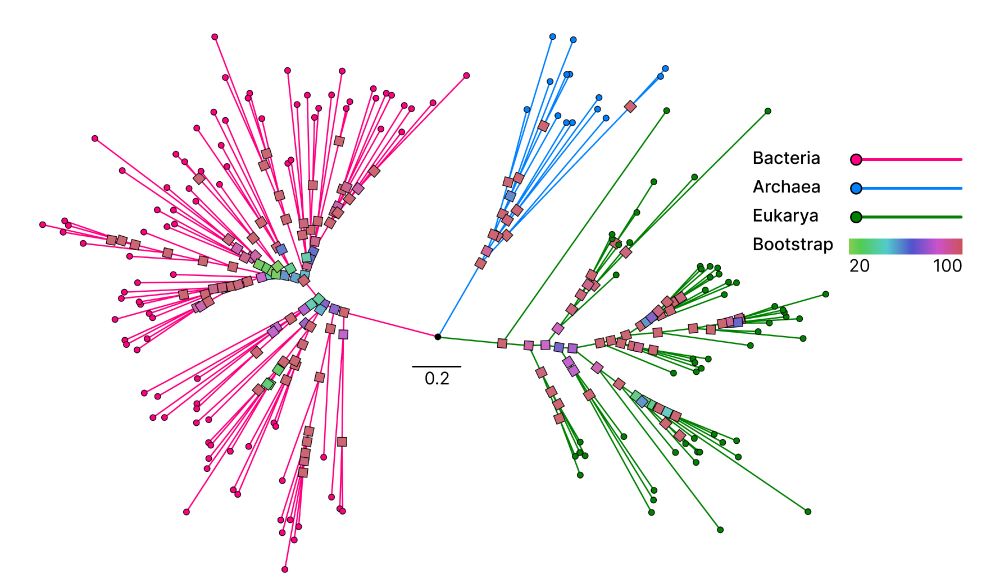
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...

