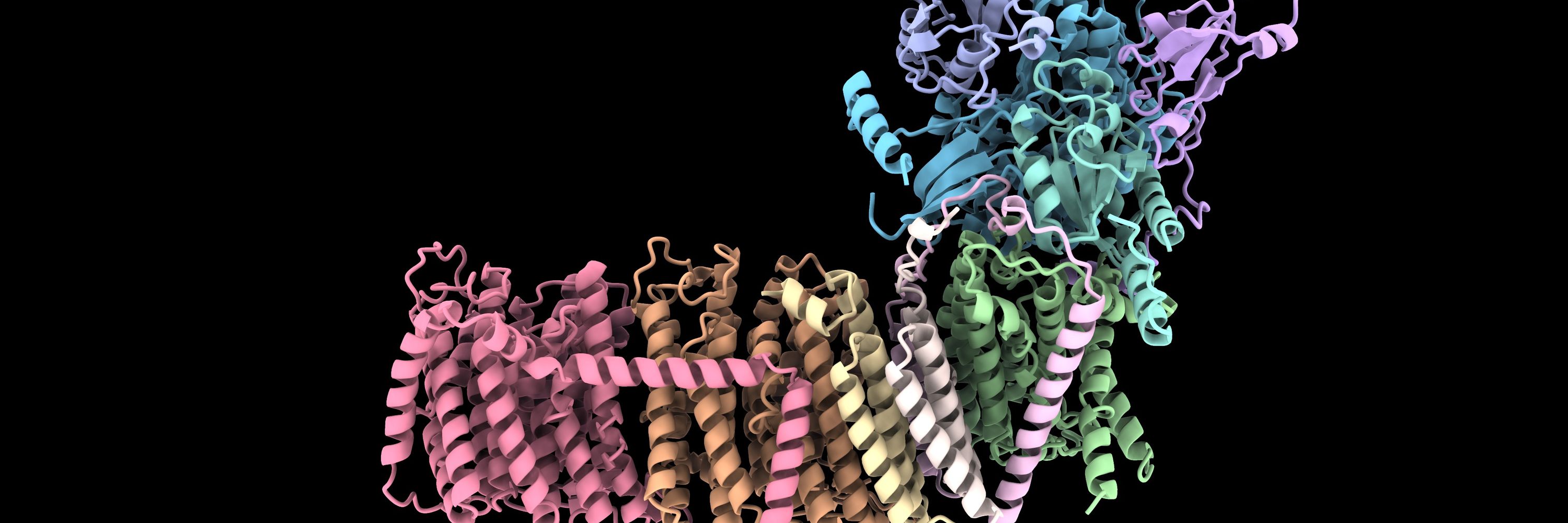
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
Links: www.nature.com/articles/s41...
www.nature.com/articles/s41...


Links: www.nature.com/articles/s41...
www.nature.com/articles/s41...


12/12

12/12


We’re diving into #cellgrowth and #lysosome biology, through the lens of molecular #structure and mechanism.
I’m thrilled to say that PhD student scholarships are available—so please share widely and get in touch if you’re interested. 🤩
We’re diving into #cellgrowth and #lysosome biology, through the lens of molecular #structure and mechanism.
I’m thrilled to say that PhD student scholarships are available—so please share widely and get in touch if you’re interested. 🤩

An important read providing some sanity against the current AI hype slop that I've been inundated with these past couple of weeks
An important read providing some sanity against the current AI hype slop that I've been inundated with these past couple of weeks




www.statnews.com/2024/06/20/r...

www.statnews.com/2024/06/20/r...





📑 Paper doi.org/10.1038/s443...
🔭 Explore virosphere viro3d.cvr.gla.ac.uk

📑 Paper doi.org/10.1038/s443...
🔭 Explore virosphere viro3d.cvr.gla.ac.uk
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations

evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations
academic.oup.com/gbe/article/...

academic.oup.com/gbe/article/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!

github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!
www.science.org/doi/10.1126/...
github.com/lehner-lab/c...

www.science.org/doi/10.1126/...
github.com/lehner-lab/c...
• it's a bad idea to install a tool on your machine that has vast read+write permissions
• code that works but is really outputting false info is a real danger and hard to catch. LLMs make that danger worse
• it's a bad idea to install a tool on your machine that has vast read+write permissions
• code that works but is really outputting false info is a real danger and hard to catch. LLMs make that danger worse
www.sciencedirect.com/science/arti... From @hkws.bsky.social

www.sciencedirect.com/science/arti... From @hkws.bsky.social
- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍

- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍

