James Lingford
@jameslingford.bsky.social
PhD student in structural biology with @greening.bsky.social and @knottrna.bsky.social at Monash Uni. (he/him)
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
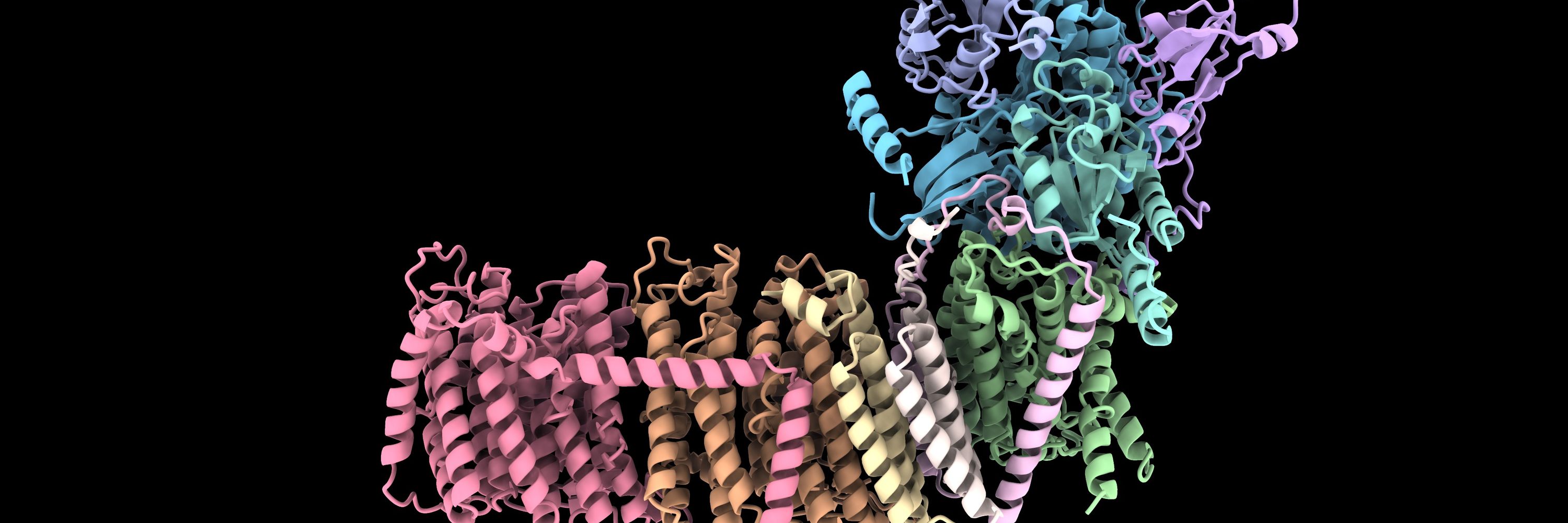
Super cool talk from @kmichie.bsky.social. Lots of examples of how AlphaFold3 does weird stuff




October 27, 2025 at 2:14 AM
Super cool talk from @kmichie.bsky.social. Lots of examples of how AlphaFold3 does weird stuff
Recent advances in the inference of deep viral
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations

September 2, 2025 at 10:35 PM
Recent advances in the inference of deep viral
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations
Got this setup working where I can now run .ipynb notebooks right from inside the terminal with a combination of neovim, quarto, kitty, and this neovim plugin called molten: github.com/benlubas/mol...
Never have to abandon my precious vim setup again
Never have to abandon my precious vim setup again
July 11, 2025 at 7:29 AM
Got this setup working where I can now run .ipynb notebooks right from inside the terminal with a combination of neovim, quarto, kitty, and this neovim plugin called molten: github.com/benlubas/mol...
Never have to abandon my precious vim setup again
Never have to abandon my precious vim setup again
Some matplotlib work in progress
June 27, 2025 at 7:13 AM
Some matplotlib work in progress
Closer... I think at this point the solution lies in manually making a list of the hex codes, but that's for another day.
June 24, 2025 at 7:58 AM
Closer... I think at this point the solution lies in manually making a list of the hex codes, but that's for another day.
Revisiting this topic now that I've forced myself to use PyMOL. Using this script to install the viridis family of colour palettes: github.com/smsaladi/pym... and running '"spectrum count, palette=magma, MODEL_NAME". The palette is not there still. They must modify the magma palette somehow

June 23, 2025 at 10:56 PM
Revisiting this topic now that I've forced myself to use PyMOL. Using this script to install the viridis family of colour palettes: github.com/smsaladi/pym... and running '"spectrum count, palette=magma, MODEL_NAME". The palette is not there still. They must modify the magma palette somehow
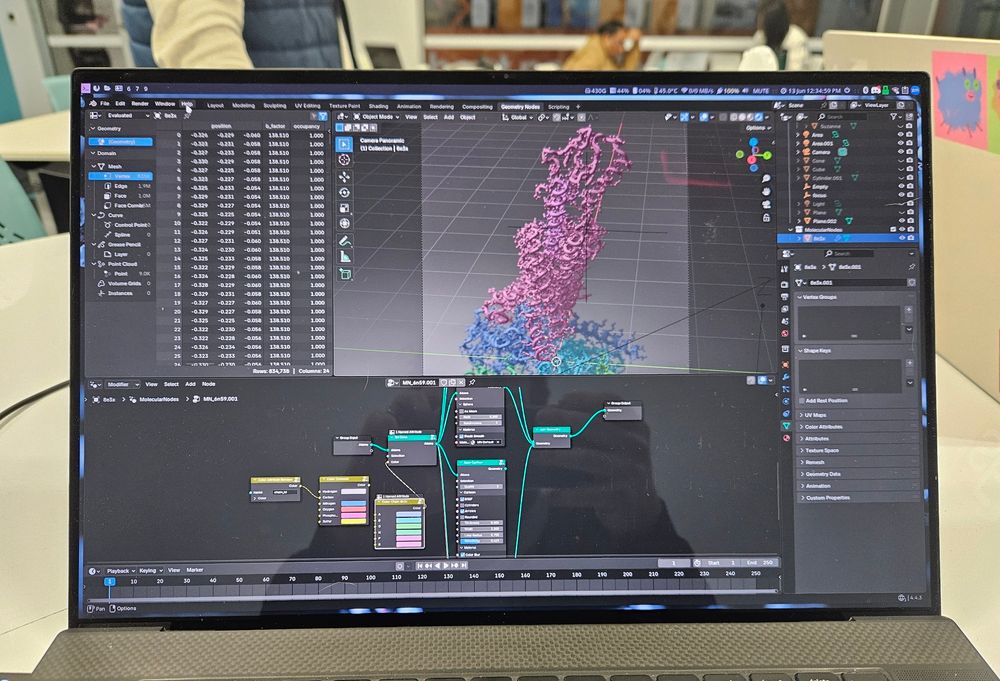
Learning some Blender molecular nodes from @sarahjpiper.bsky.social @ccemmp-outreach.bsky.social
June 13, 2025 at 4:03 AM
Learning some Blender molecular nodes from @sarahjpiper.bsky.social @ccemmp-outreach.bsky.social
Little shell scripting solution to the problem of finding a group A in column1 of a table that is entirely made up of group B in column2, and sorting for the biggest homogeneous grouping.
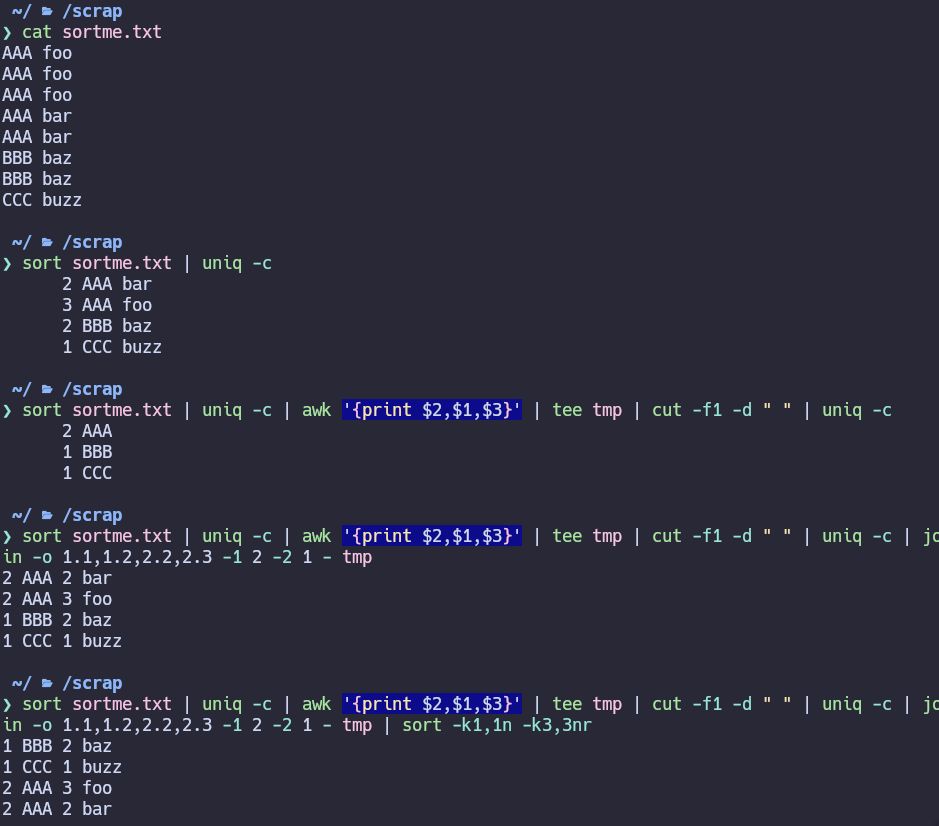
June 11, 2025 at 1:49 AM
Little shell scripting solution to the problem of finding a group A in column1 of a table that is entirely made up of group B in column2, and sorting for the biggest homogeneous grouping.
I'm sick of typing "conda activate ENV" or "conda deactivate" to switch virtual environments. Now I've added a conda/mamba env switcher function that uses fzf to my ~/.zshrc.
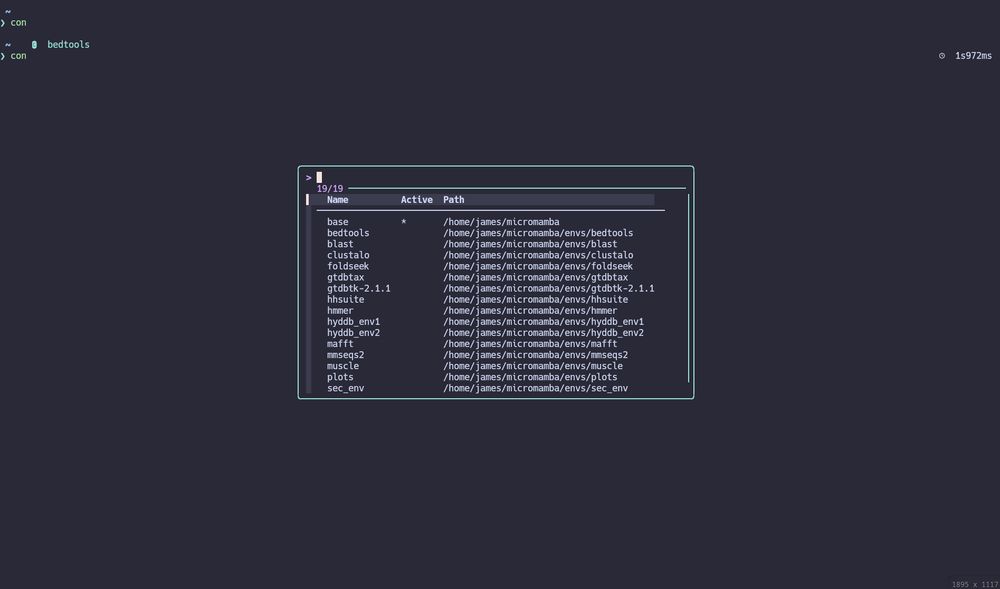
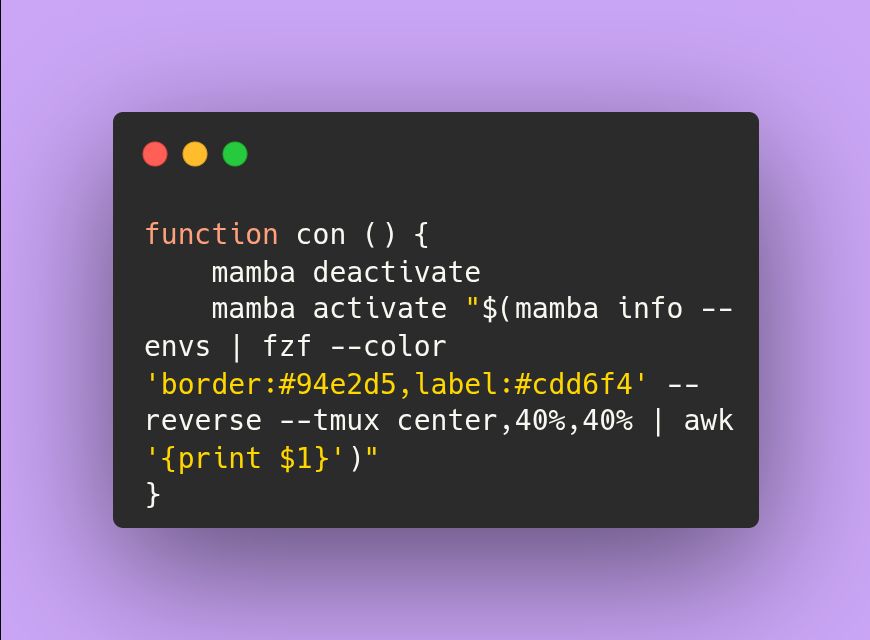
June 3, 2025 at 7:12 AM
I'm sick of typing "conda activate ENV" or "conda deactivate" to switch virtual environments. Now I've added a conda/mamba env switcher function that uses fzf to my ~/.zshrc.
Awk command to unwrap wrapped lines in fasta files. It's just neat!
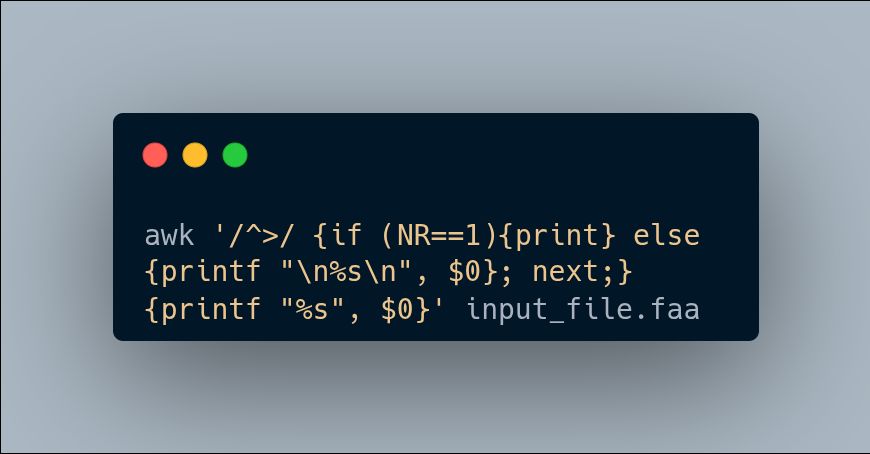
May 18, 2025 at 11:47 AM
Awk command to unwrap wrapped lines in fasta files. It's just neat!
Bioenergetics and the evolution of cellular traits [review]:
doi.org/10.1146/annu...
doi.org/10.1146/annu...
![The energetic cost of cellular traits. Costs were calculated for individual proteins, for macromolecular
complexes, or, in the case of Escherichia coli, for a single cell (shown for comparison). Calmodulin: 148 amino acids; ATP synthase: 4,955 amino acids (E. coli); dynein-2 motor: 12,084 amino acids (human, PDB ID:
6SC2); ribosome: 7,459 amino acids and 4,566 ribonucleotides (E. coli); vesicle: diameter 50 nm [membrane cost including proteins D 1.45 109
ATP mm 2
(86)]; nuclear pore complexes: 50–110 MDa (yeast and
vertebrates) (2, 7); eukaryotic flagellum: 11 mm in length; and E. coli: estimated using the fourth cell budget method. Abbreviation: PDB ID, Protein Data Bank identifier.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:arnopirhmzvadspoff2l3235/bafkreigbbrxtwjtzwpok56eq5qrizpts43xlmts5z2wxlabtvzi7nrk5n4@jpeg)
May 10, 2025 at 5:05 AM
Bioenergetics and the evolution of cellular traits [review]:
doi.org/10.1146/annu...
doi.org/10.1146/annu...
Reading old papers is cool because you never see this sort of casual tone anymore. www.sciencedirect.com/science/arti...
![Not surprisingly, Fearnley and Walker [30]
could confirm the sequence relationships with the
ORF7 gene of the E. coli formate-hydrogen-lyase sys-
tem. Once I realized the possible implications of these
links, it was only logical to compare the sequence of
the PSST gene [29] with those of the small subunits of
nickel hydrogenases. The result is summarized in Fig. 1.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:arnopirhmzvadspoff2l3235/bafkreihdulsfcudwfdq5f3eucgri2rdikf2srwilk4blozkmhyzwvzvj3i@jpeg)

April 28, 2025 at 6:11 AM
Reading old papers is cool because you never see this sort of casual tone anymore. www.sciencedirect.com/science/arti...
ChimeraX tip: to get a lined border around an internal surface requires superposing two separate copies of your model. One for showing the surrounding cartoon structure, and the other showing just the surface (and select that one to give it a border)
www.jameslingford.com/blog/chimera...
www.jameslingford.com/blog/chimera...

April 19, 2025 at 6:56 AM
ChimeraX tip: to get a lined border around an internal surface requires superposing two separate copies of your model. One for showing the surrounding cartoon structure, and the other showing just the surface (and select that one to give it a border)
www.jameslingford.com/blog/chimera...
www.jameslingford.com/blog/chimera...
A modified viridis palette looks pretty nice too.

April 17, 2025 at 11:00 AM
A modified viridis palette looks pretty nice too.
Made my terminal colours pretty, so now I'm a true hacker 😎

April 17, 2025 at 10:52 AM
Made my terminal colours pretty, so now I'm a true hacker 😎
Alright, this might be as good as it gets:
rainbow palette magma; col modify blackness - 70; col modify whiteness - 80; col modify saturation - 70; col modify lightness + 22;
Increasing the lightness recreates the washed out transparent look without touching the transparency setting
rainbow palette magma; col modify blackness - 70; col modify whiteness - 80; col modify saturation - 70; col modify lightness + 22;
Increasing the lightness recreates the washed out transparent look without touching the transparency setting

April 16, 2025 at 8:12 AM
Alright, this might be as good as it gets:
rainbow palette magma; col modify blackness - 70; col modify whiteness - 80; col modify saturation - 70; col modify lightness + 22;
Increasing the lightness recreates the washed out transparent look without touching the transparency setting
rainbow palette magma; col modify blackness - 70; col modify whiteness - 80; col modify saturation - 70; col modify lightness + 22;
Increasing the lightness recreates the washed out transparent look without touching the transparency setting
Close to recreating it in ChimeraX:
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation

April 16, 2025 at 6:31 AM
Close to recreating it in ChimeraX:
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation
1. Install the python file: github.com/smsaladi/chi...
2. rainbow palette magma
3. col modify #1 blackness - 40
4. col modify #1 whiteness - 40
5. col modify #1 saturation - 50
6. col modify #1 lightness + 20
Room for improvement, needs more desaturation
Something I've been wondering is what sort of settings the Baker lab is using to get these gorgeous PyMOL figures with? (I'm assuming it's PyMOL?)

April 16, 2025 at 5:25 AM
Something I've been wondering is what sort of settings the Baker lab is using to get these gorgeous PyMOL figures with? (I'm assuming it's PyMOL?)
Crash course in MD-sim today 💻
April 11, 2025 at 5:09 AM
Crash course in MD-sim today 💻
Screwed up my Linux machine by trying to be clever. So here we go again with a fresh install

April 7, 2025 at 1:06 AM
Screwed up my Linux machine by trying to be clever. So here we go again with a fresh install
Hydrogenase-driven ATP synthesis from air
www.biorxiv.org/content/10.1...
New work from us where we put the Huc hydrogenase to work to see if it can generate a proton motive force. It can!
cc: @ashleighkropp.bsky.social @rhyswg.bsky.social @greening.bsky.social
www.biorxiv.org/content/10.1...
New work from us where we put the Huc hydrogenase to work to see if it can generate a proton motive force. It can!
cc: @ashleighkropp.bsky.social @rhyswg.bsky.social @greening.bsky.social
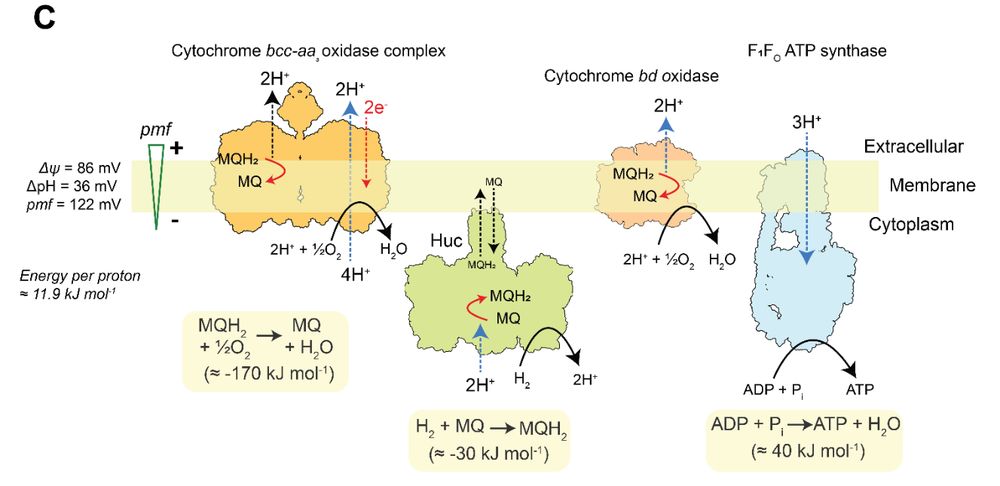
March 19, 2025 at 3:21 AM
Hydrogenase-driven ATP synthesis from air
www.biorxiv.org/content/10.1...
New work from us where we put the Huc hydrogenase to work to see if it can generate a proton motive force. It can!
cc: @ashleighkropp.bsky.social @rhyswg.bsky.social @greening.bsky.social
www.biorxiv.org/content/10.1...
New work from us where we put the Huc hydrogenase to work to see if it can generate a proton motive force. It can!
cc: @ashleighkropp.bsky.social @rhyswg.bsky.social @greening.bsky.social
Goodbye @lorneproteins.bsky.social 2025, and thanks for the great conference. I'll miss getting to see hundreds of cockatoos whenever I step outside

February 12, 2025 at 9:59 PM
Goodbye @lorneproteins.bsky.social 2025, and thanks for the great conference. I'll miss getting to see hundreds of cockatoos whenever I step outside
Finally got around to reading the FoldMason paper for structural phylogenetics. Fantastic work
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

January 13, 2025 at 9:09 AM
Finally got around to reading the FoldMason paper for structural phylogenetics. Fantastic work
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

