GPCRs | synthetic biology | genomics
Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social

A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social
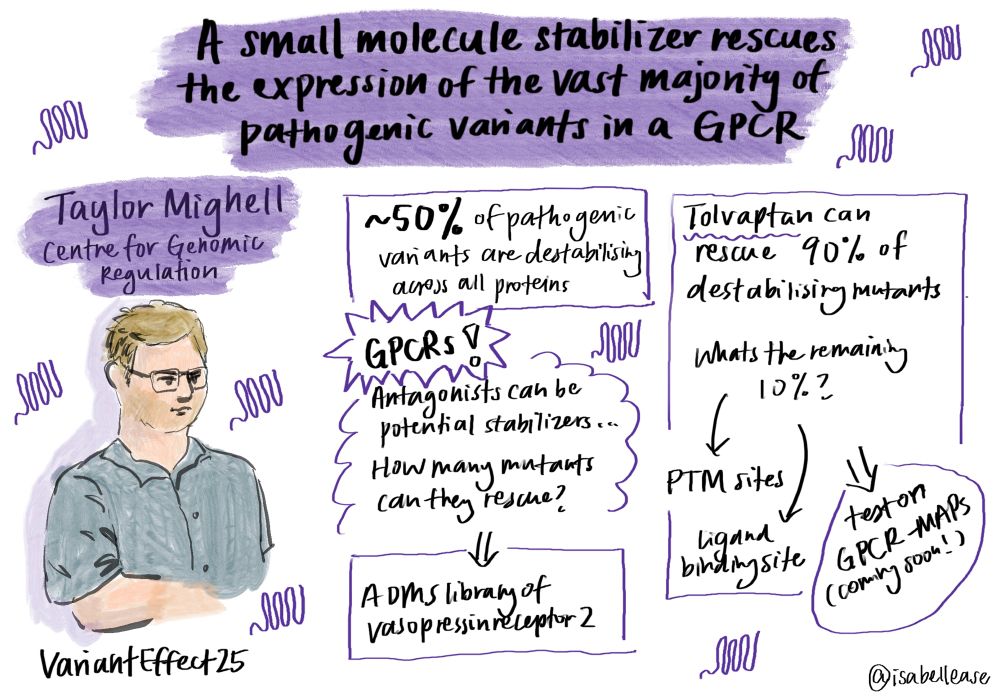
@taylor-mighell.bsky.social from @benlehner.bsky.social lab presents: Two wrongs make a right: can GPCR antagonists rescue mutant receptors? 🧬🧪🔁

@taylor-mighell.bsky.social from @benlehner.bsky.social lab presents: Two wrongs make a right: can GPCR antagonists rescue mutant receptors? 🧬🧪🔁