doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
Together with Christine Moene @cmoene.bsky.social, we explored what happens when you scramble the genome—revealing how Sox2’s position shapes enhancer activation.
📖 Read the full story here: www.science.org/doi/10.1126/...

Together with Christine Moene @cmoene.bsky.social, we explored what happens when you scramble the genome—revealing how Sox2’s position shapes enhancer activation.
📖 Read the full story here: www.science.org/doi/10.1126/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
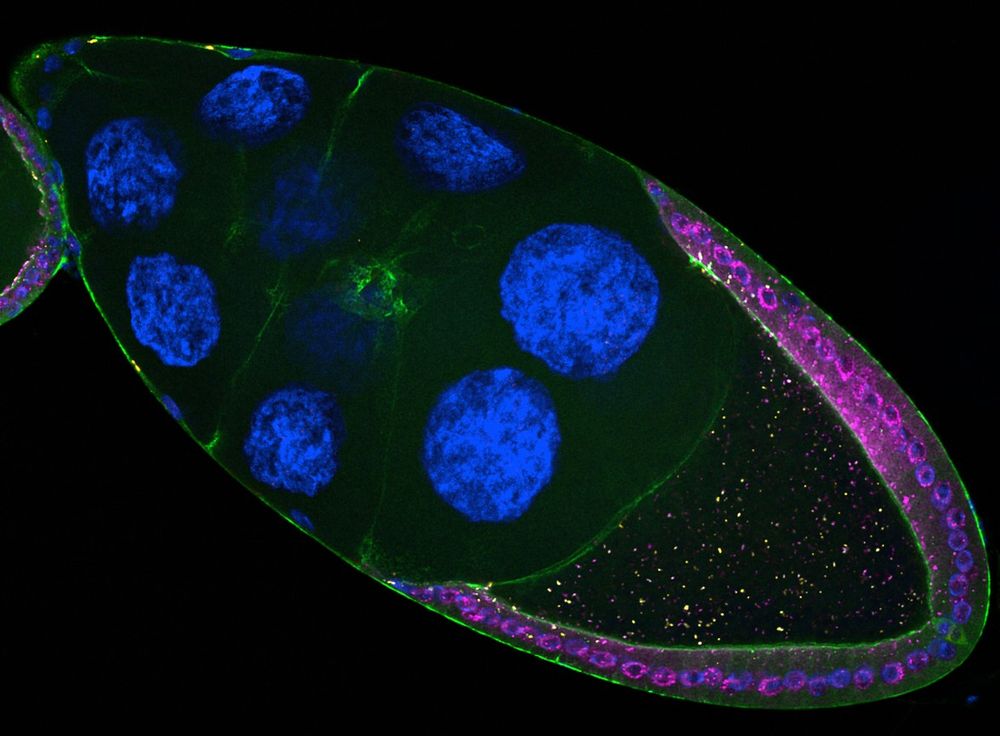
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
www.annualreviews.org/content/jour...
www.annualreviews.org/content/jour...
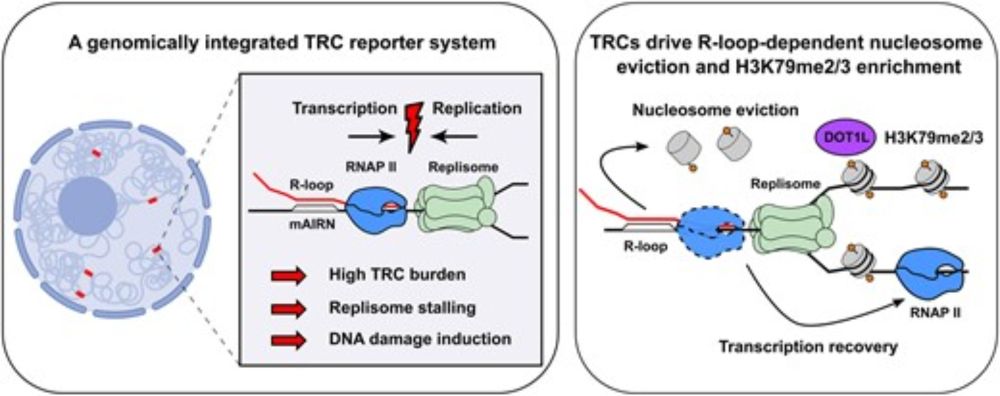
academic.oup.com/nar/article/...
#TRC #r-loop #chromatin
More below 👇
academic.oup.com/nar/article/...
#TRC #r-loop #chromatin
More below 👇

