
Professor of Genetics, Strasbourg University
www.cell.com/cell-genomic...

www.cell.com/cell-genomic...
genetics-gsa.org/yeast-2026/

genetics-gsa.org/yeast-2026/
www.nature.com/articles/d41...
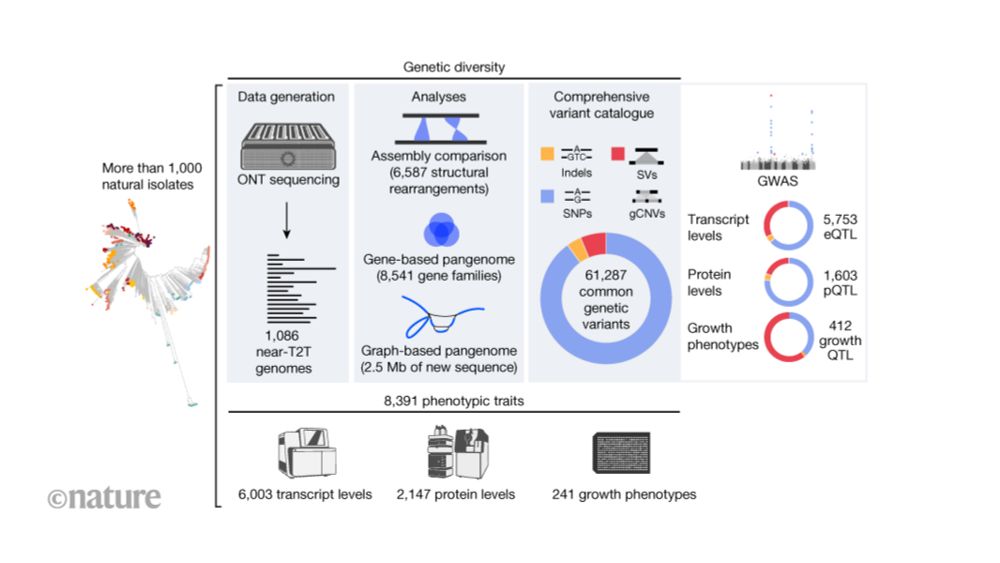
www.nature.com/articles/d41...




go.nature.com/4nVsl9z

go.nature.com/4nVsl9z
This work has been a major project of @tbadet.bsky.social during his time in our lab.
We hope to stimulate much more exciting work on fungal TEs.
journals.plos.org/plosbiology/...
This work has been a major project of @tbadet.bsky.social during his time in our lab.
We hope to stimulate much more exciting work on fungal TEs.
journals.plos.org/plosbiology/...
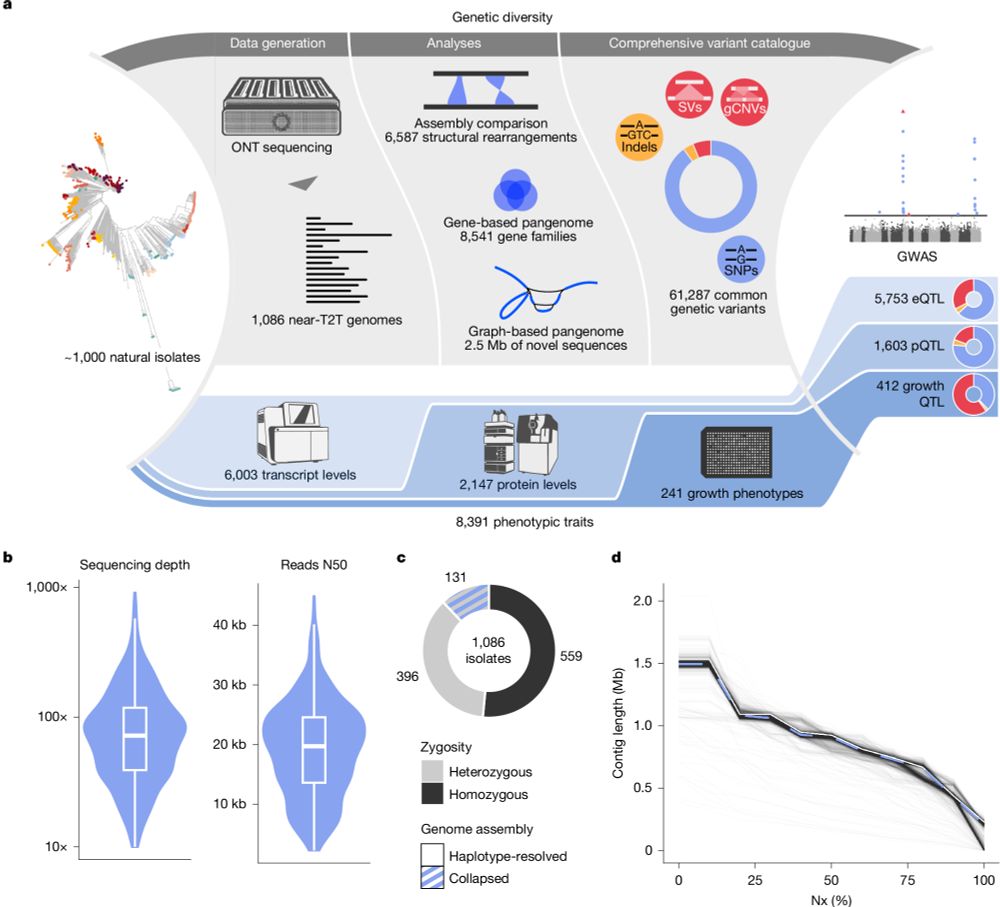
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Browse abstract submission topics, key dates, speaker & venue info, details on travel funding, and more: buff.ly/HFIt31b
Bookmark the page to stay updated as new information becomes available!

Browse abstract submission topics, key dates, speaker & venue info, details on travel funding, and more: buff.ly/HFIt31b
Bookmark the page to stay updated as new information becomes available!

Join at the GSA Yeast 2026 meeting in beautiful Asilomar, Pacific Grove, CA!
📅 Save the date: June 14–18, 2026
➡️ genetics-gsa.org/yeast-2026/

Join at the GSA Yeast 2026 meeting in beautiful Asilomar, Pacific Grove, CA!
📅 Save the date: June 14–18, 2026
➡️ genetics-gsa.org/yeast-2026/
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.surveymonkey.com/r/Yeast26?ut...

www.surveymonkey.com/r/Yeast26?ut...
www.surveymonkey.com/r/Yeast26?ut...

www.surveymonkey.com/r/Yeast26?ut...
www.nature.com/articles/s41...

www.nature.com/articles/s41...




cordis.europa.eu/article/id/4...

cordis.europa.eu/article/id/4...
@unistra.fr @cnrsbiologie.bsky.social
www.nature.com/articles/s41...
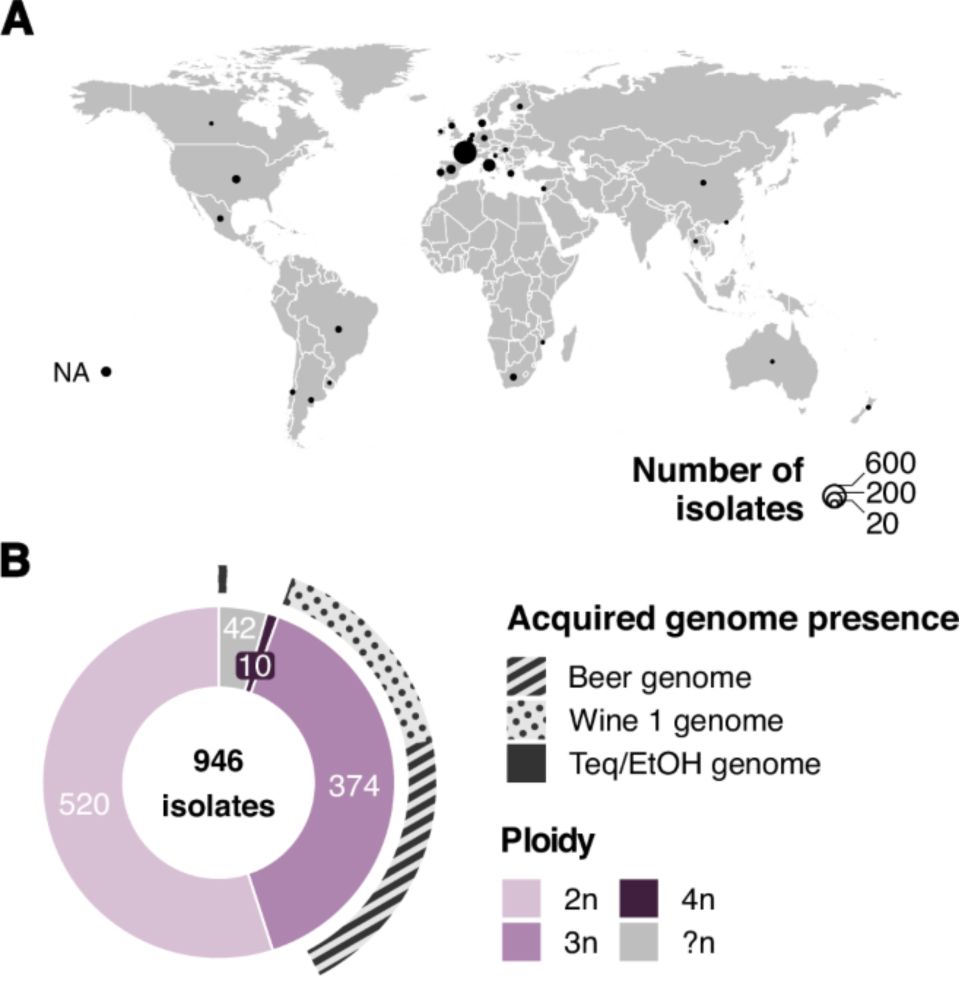
@unistra.fr @cnrsbiologie.bsky.social
www.nature.com/articles/s41...

@unistra.fr @cnrsbiologie.bsky.social
www.nature.com/articles/s41...
@unistra.fr @cnrsbiologie.bsky.social #IUF
bsky.app/profile/erc....
The new funding, worth nearly €721 million, is part of the 🇪🇺 EU’s #HorizonEurope programme.
Find out more 👉 europa.eu/!JYKxkD
#ERCAdG #FrontierResearch @ec.europa.eu
@unistra.fr @cnrsbiologie.bsky.social #IUF
bsky.app/profile/erc....
www.nature.com/articles/s41...
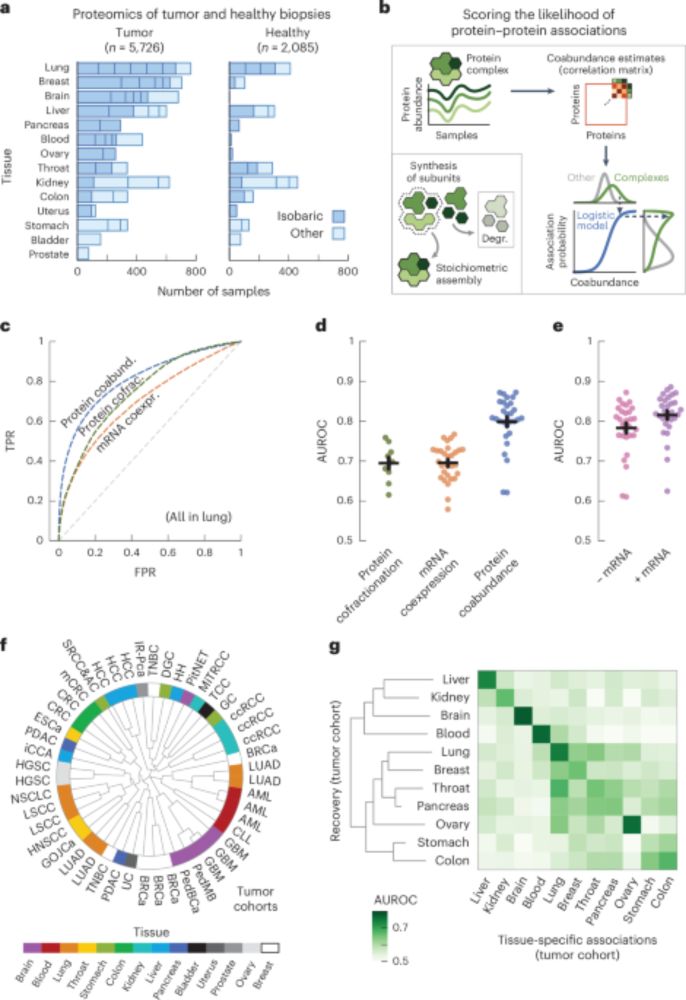
www.nature.com/articles/s41...
🔹Opening remarks by Flora Vincent 🙌🏼
🔹The first talk of the day: 'Unlocking nature’s arsenal: investigating phage-bacteria dynamics in aquatic ecosystems for sustainable aquaculture' presented by Frédérique Le Roux




🔹Opening remarks by Flora Vincent 🙌🏼
🔹The first talk of the day: 'Unlocking nature’s arsenal: investigating phage-bacteria dynamics in aquatic ecosystems for sustainable aquaculture' presented by Frédérique Le Roux

