Interested in dynamic effects of genetic variation across differentiation
(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com

(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com

www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...

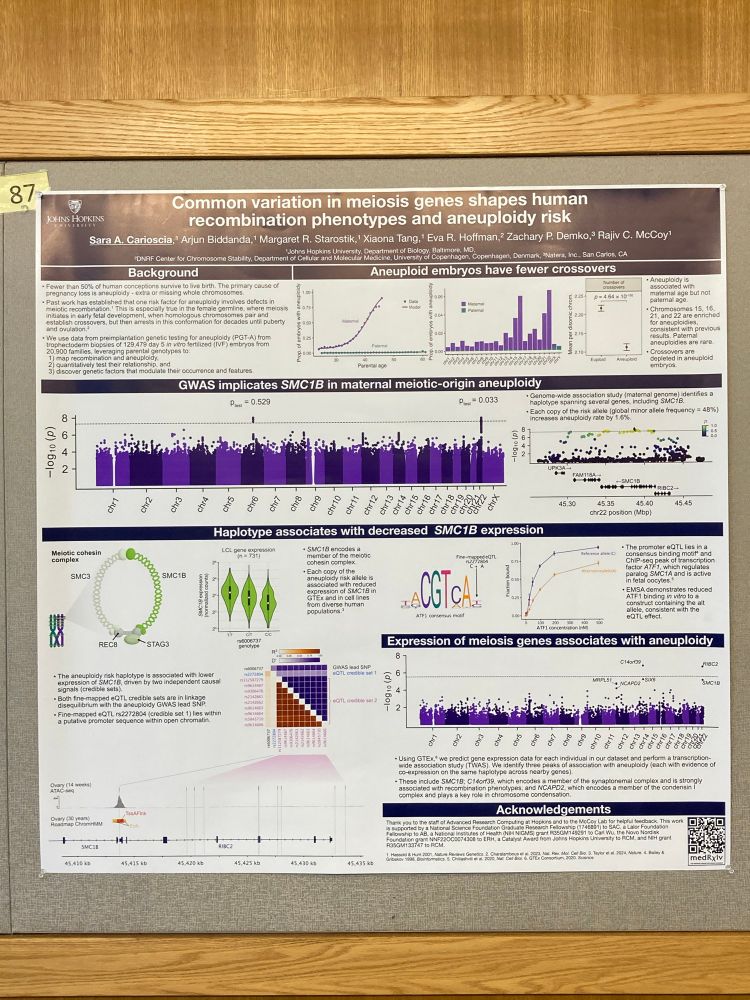
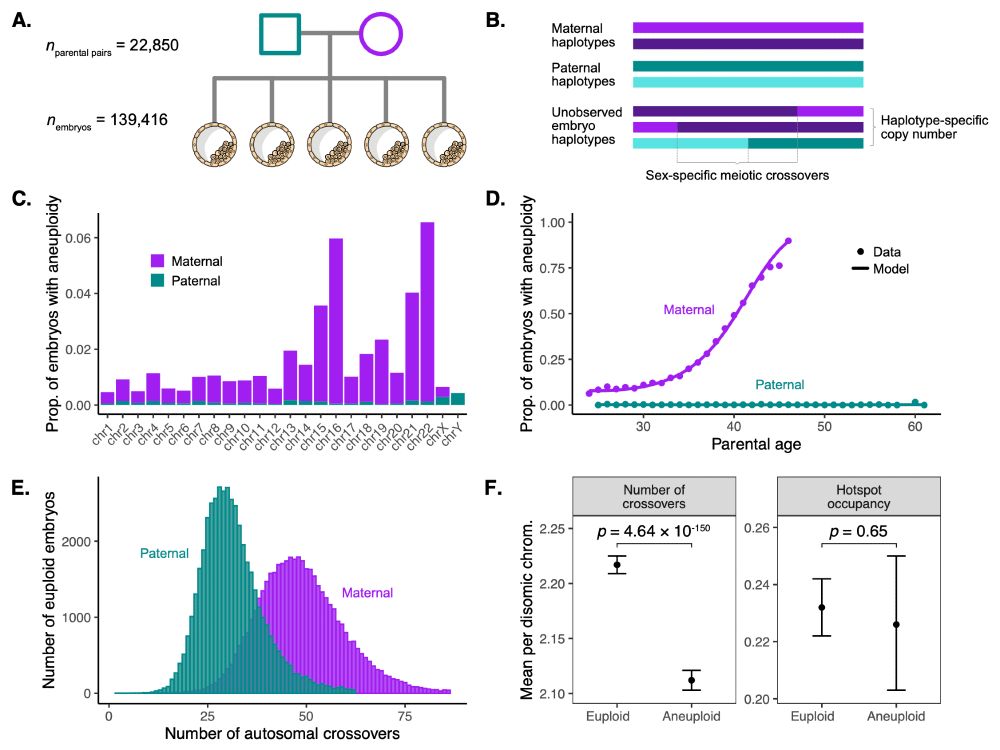
(For the record, no one has ever asked me this, but it is a really good question!)
I think we know where they are.
(For the record, no one has ever asked me this, but it is a really good question!)
I think we know where they are.
It’s available on Amazon: tinyurl.com/mx2hewen

It’s available on Amazon: tinyurl.com/mx2hewen


Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
To understand gene regulation across diverse environmental conditions and cellular contexts, we treated a broad array of human cell types with three environmental exposures in vitro.
www.biorxiv.org/content/10.1...
You can find out more about me at these links:
LinkedIn: www.linkedin.com/in/arun96/
Personal Website: arundas.org
I’m going to be defending my thesis on Wednesday, so I thought this was as good a time as any to introduce myself and my work.
I’m Arun Das, I’m a PhD student in Schatz Lab @ JHU, and my work broadly focuses on algorithms to improve accessibility and representation in genomics.
I’m going to be defending my thesis on Wednesday, so I thought this was as good a time as any to introduce myself and my work.
I’m Arun Das, I’m a PhD student in Schatz Lab @ JHU, and my work broadly focuses on algorithms to improve accessibility and representation in genomics.
www.medrxiv.org/content/10.1...
www.medrxiv.org/content/10.1...
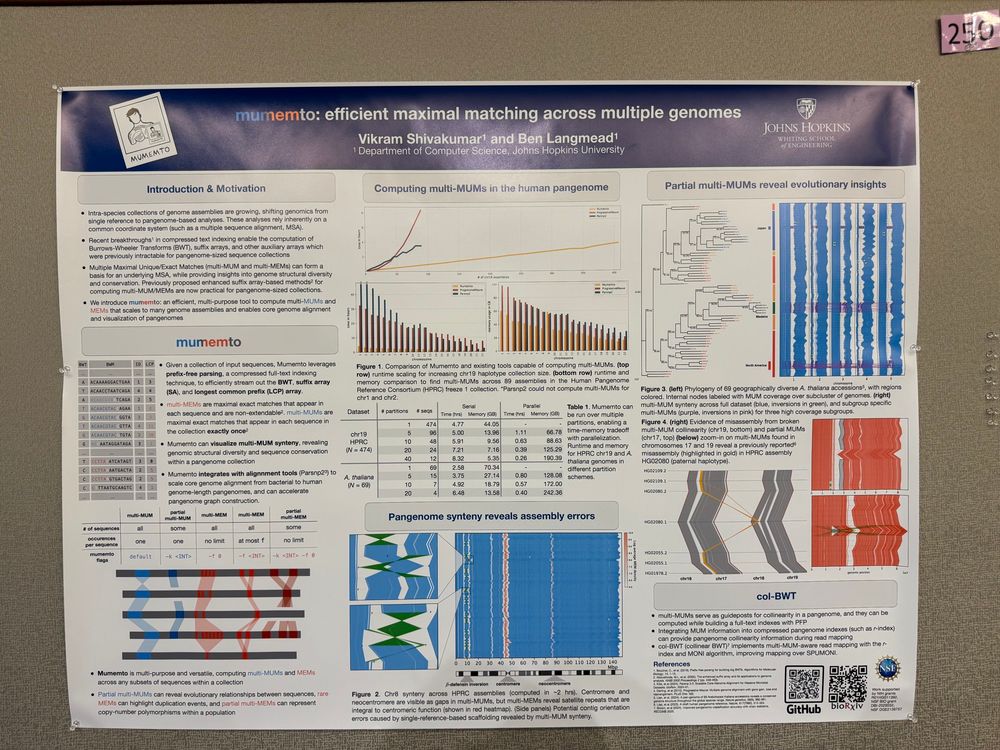
Github: github.com/vikshiv/mume...
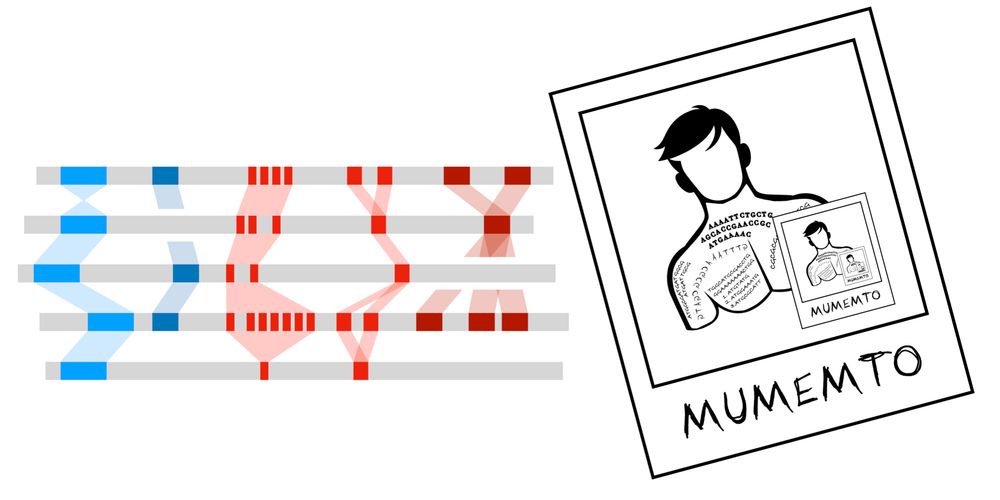
Github: github.com/vikshiv/mume...



