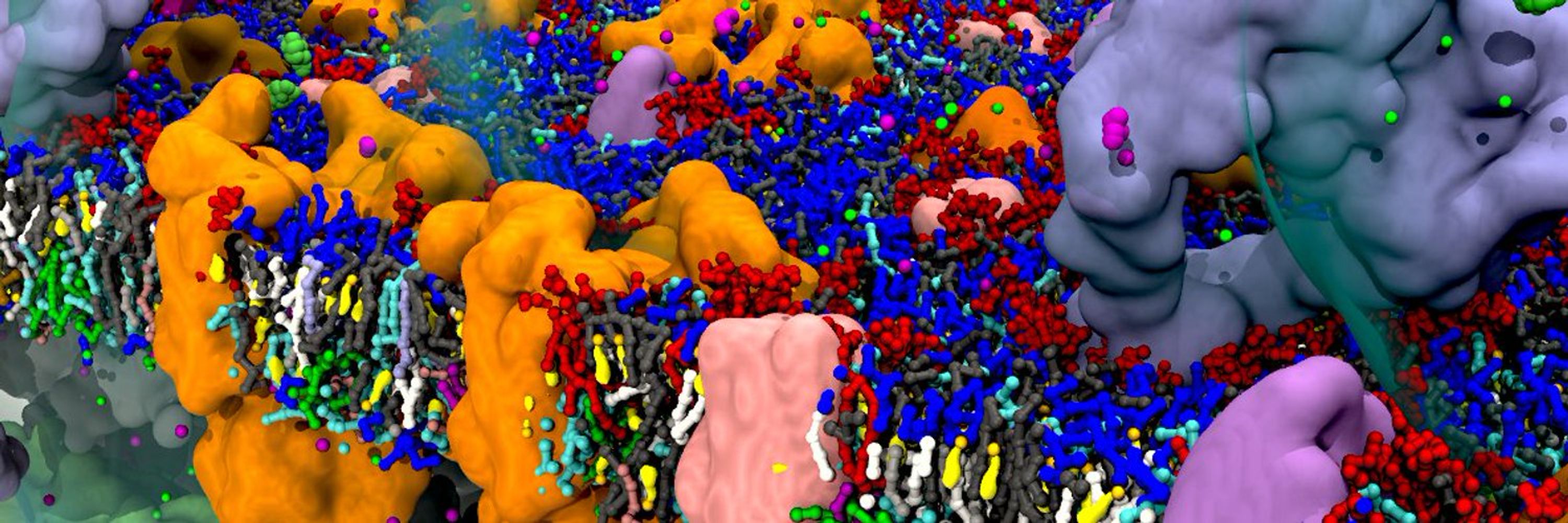
@acscomp.bsky.social reception Sunday as well. 😄🍸
@acscomp.bsky.social reception Sunday as well. 😄🍸
📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry

📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
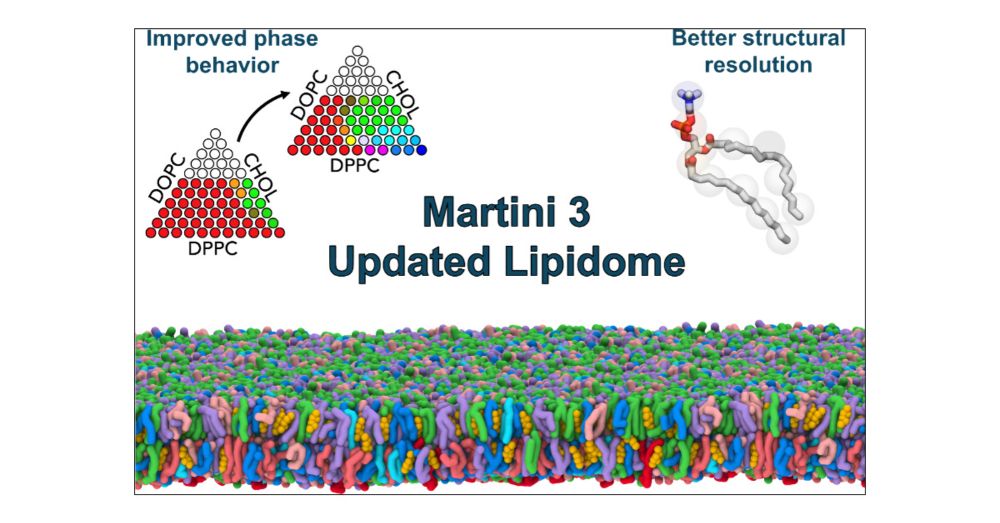
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
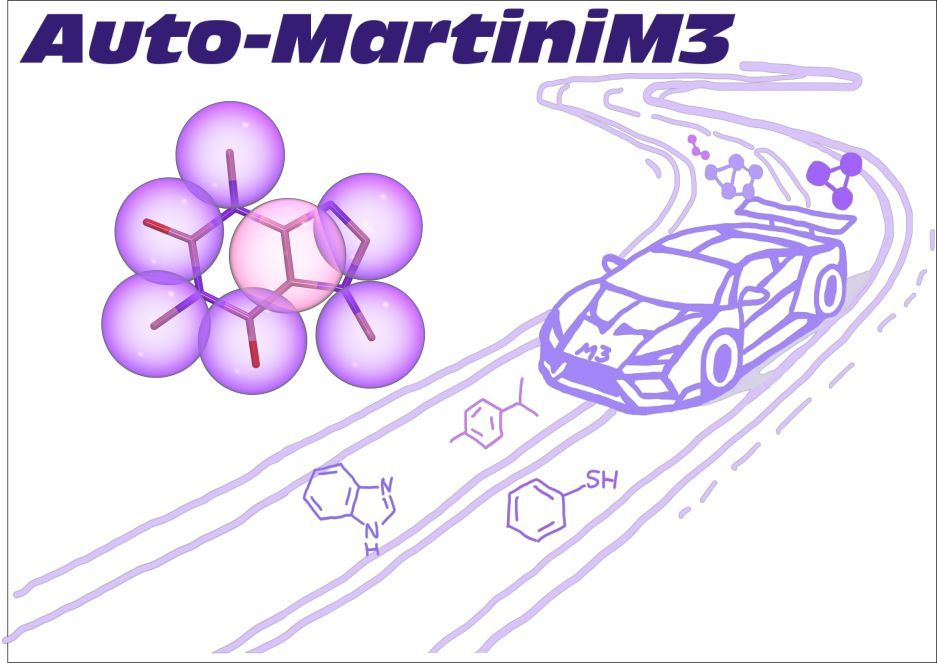
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
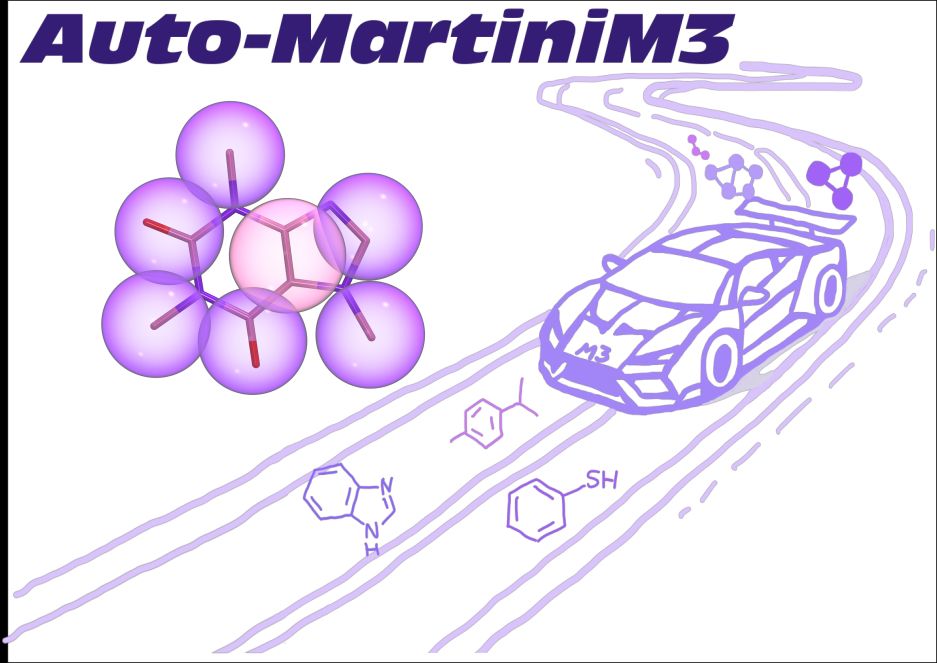
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social

We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...

We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social

🔗 Read here: doi.org/10.1021/acs....

🔗 Read here: doi.org/10.1021/acs....
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
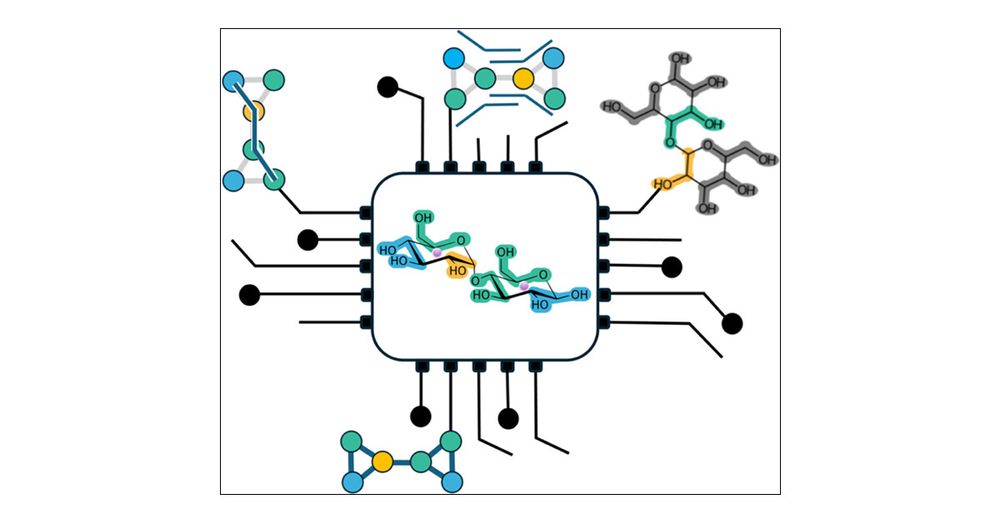
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation

We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
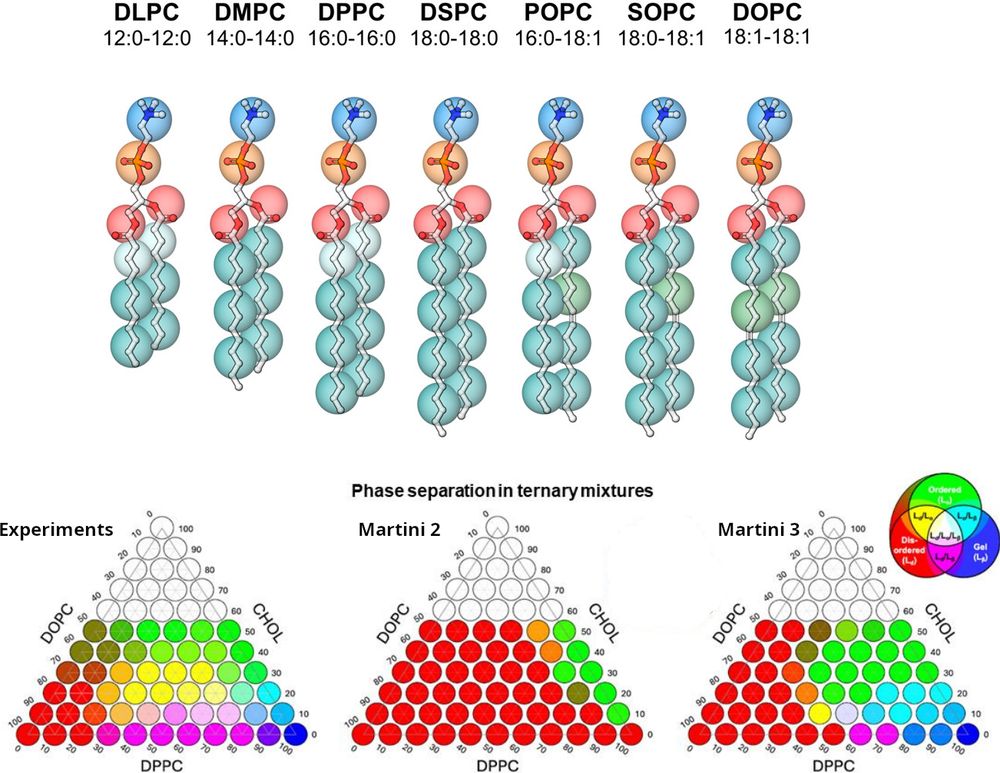
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
#CompChem
chemrxiv.org/engage/chemr...

#CompChem
chemrxiv.org/engage/chemr...
Read the full article here: rupress.org/jcb/article/...

Read the full article here: rupress.org/jcb/article/...

