Paulo C. T. Souza
@pauloctsouza.bsky.social
Martini developer trying to make CG models more useful for molecular design and drug/vaccine delivery 🖥️+🍸=💊💉 | Group leader at CBPsmn/ENS de Lyon | CNRS Researcher at DAMM team of LBMC/ENS de Lyon | Scientific consultant
Looking forward to #acsfall2025! Hoping to finally meet in person the colleagues I’ve long admired at the
@acscomp.bsky.social reception Sunday as well. 😄🍸
@acscomp.bsky.social reception Sunday as well. 😄🍸
August 14, 2025 at 9:35 PM
Looking forward to #acsfall2025! Hoping to finally meet in person the colleagues I’ve long admired at the
@acscomp.bsky.social reception Sunday as well. 😄🍸
@acscomp.bsky.social reception Sunday as well. 😄🍸
🔬 Coarse-Grained Modeling @ ACS Fall 2025 — Program Out Now! 🚀
📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry
📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry

August 13, 2025 at 10:43 PM
🔬 Coarse-Grained Modeling @ ACS Fall 2025 — Program Out Now! 🚀
📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry
📍 Washington DC | Aug 17–21
- Wed, Aug 20 AM: LNPs — nucleic acid delivery → viral entry simulations
- Wed, Aug 20 PM: Proteins & multiscale modeling
- Thu, Aug 21 AM: Methods & industry
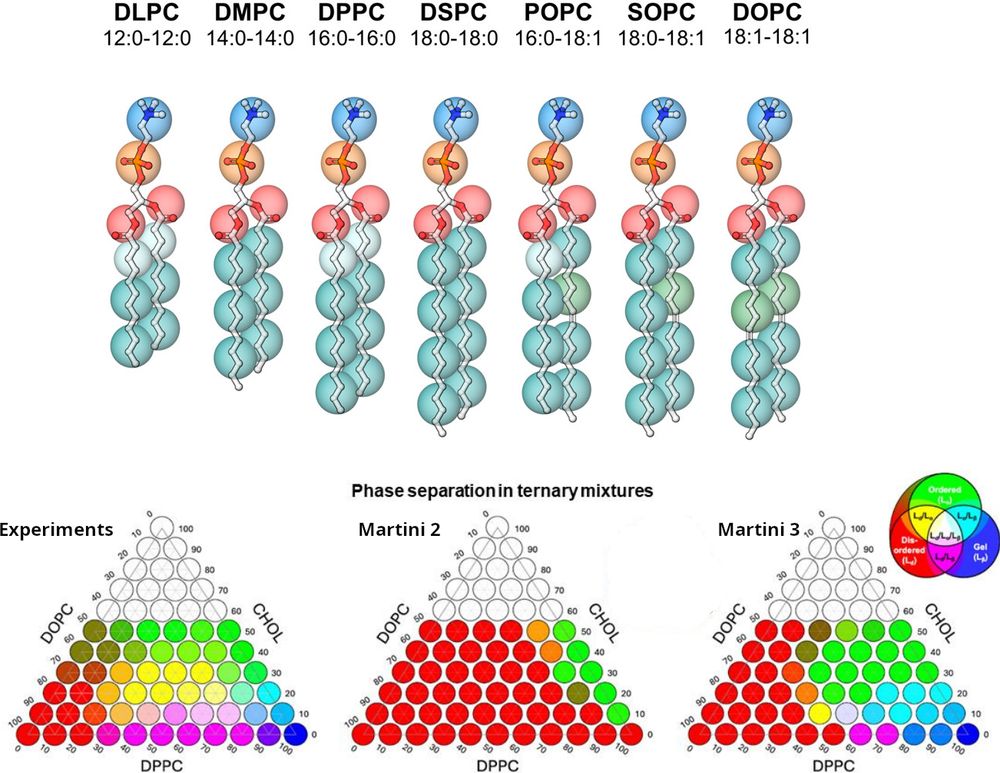
Our collaborative work "The #Martini3 #Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior" is now published in #ACSCentralScience! 🎉
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
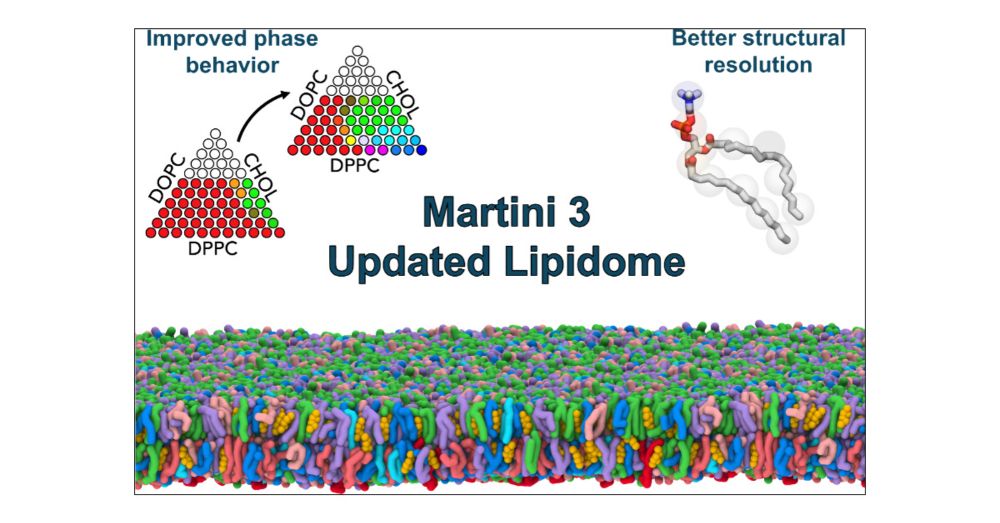
The Martini 3 Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior
Lipid membranes are central to cellular life. Complementing experiments, computational modeling has been essential in unraveling complex lipid-biomolecule interactions, crucial in both academia and industry. The Martini model, a coarse-grained force field for efficient molecular dynamics simulations, is widely used to study membrane phenomena but has faced limitations, particularly in capturing realistic lipid phase behavior. Here, we present refined Martini 3 lipid models with a mapping scheme that distinguishes lipid tails that differ by just two carbon atoms, enhancing the structural resolution and thermodynamic accuracy of model membrane systems including ternary mixtures. The expanded Martini lipid library includes thousands of models, enabling simulations of complex and biologically relevant systems. These advancements establish Martini as a robust platform for lipid-based simulations across diverse fields.
pubs.acs.org
August 1, 2025 at 1:49 PM
Our collaborative work "The #Martini3 #Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior" is now published in #ACSCentralScience! 🎉
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
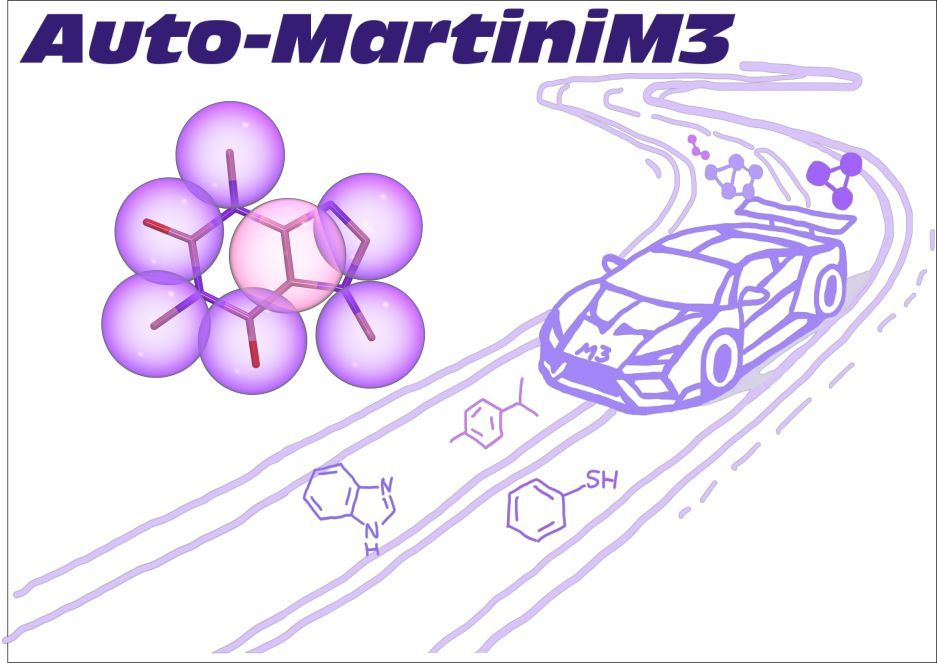
🚀 New preprint out!
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
July 18, 2025 at 10:12 PM
🚀 New preprint out!
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
"Fast Parameterization of #Martini3 Models for Fragments and Small Molecules" is now on #bioRxiv
👉 doi.org/10.1101/2025...
#AutoMartiniM3 – tool for automated #CG modeling. With @matthchavent.bsky.social, @tbereau.bsky.social and others.
#CoarseGraining #MD #DrugDiscovery
Reposted by Paulo C. T. Souza
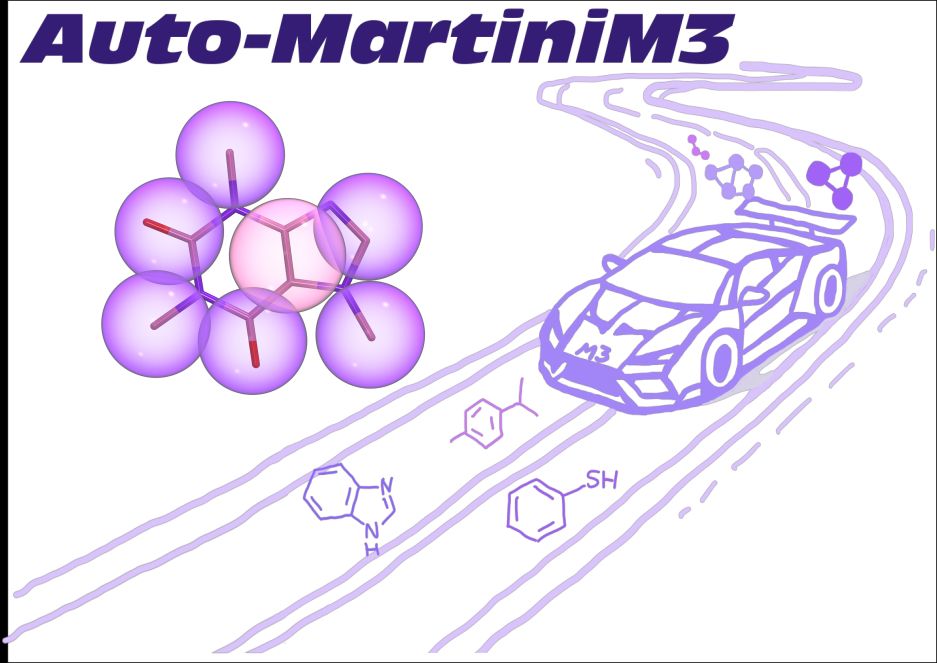
Very happy to release AutoMartiniM3 for Martini3 to automatize CG modelling of small molecules and fragments.
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
June 11, 2025 at 6:49 AM
Very happy to release AutoMartiniM3 for Martini3 to automatize CG modelling of small molecules and fragments.
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
Great work from M Szczuka in my lab in collab with @pauloctsouza.bsky.social and @tbereau.bsky.social teams!
Give it a try: github.com/Martini-Forc...
poke @cg-martini.bsky.social
Reposted by Paulo C. T. Souza

June 7, 2025 at 4:15 PM
New paper out in Nature Communications from @natureportfolio.nature.com :
We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...
We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...

GōMartini 3: From large conformational changes in proteins to environmental bias corrections - Nature Communications
This work presents GōMartini 3, an improved coarse-grained protein model combining physics- and structure-based approaches. It boosts computational efficiency and accuracy for structured soluble and m...
www.nature.com
April 30, 2025 at 10:03 PM
New paper out in Nature Communications from @natureportfolio.nature.com :
We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...
We present #GōMartini 3 — combining structure-based Gōwith #Martini3 to better capture #protein conformational changes and environmental effects.
📄 www.nature.com/articles/s41...
Reposted by Paulo C. T. Souza
🔬Just published in Bioinformatics Advances: "Challenges in predicting PROTAC-mediated protein-protein interfaces with AlphaFold reveal a general limitation on small interfaces": https://doi.org/10.1093/bioadv/vbaf056
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social
doi.org
March 27, 2025 at 12:03 PM
🔬Just published in Bioinformatics Advances: "Challenges in predicting PROTAC-mediated protein-protein interfaces with AlphaFold reveal a general limitation on small interfaces": https://doi.org/10.1093/bioadv/vbaf056
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social
#ProteinDesign #AlphaFold #StructuralBiology
@pauloctsouza.bsky.social
🎉 Big news! 🎉 Together with Jianing Li (Purdue Univ.) & John Shelley (@schrodingerinc.bsky.social), I’m organizing a symposium on #CG #Modeling for Molecular & Formulation Design at ACS Fall 2025 (Aug 17-21, Washington, DC)! 🔬✨ For my part, I am grateful for the support of @ensdelyon.bsky.social!

February 4, 2025 at 9:02 PM
🎉 Big news! 🎉 Together with Jianing Li (Purdue Univ.) & John Shelley (@schrodingerinc.bsky.social), I’m organizing a symposium on #CG #Modeling for Molecular & Formulation Design at ACS Fall 2025 (Aug 17-21, Washington, DC)! 🔬✨ For my part, I am grateful for the support of @ensdelyon.bsky.social!
🚀 Check out our latest review, "Computational Methods for Modeling Lipid-Mediated Active Pharmaceutical Ingredient Delivery", now published in Molecular Pharmaceutics (@ACS Journals)! 🧪📖
🔗 Read here: doi.org/10.1021/acs....
🔗 Read here: doi.org/10.1021/acs....

Computational Methods for Modeling Lipid-Mediated Active Pharmaceutical Ingredient Delivery
Lipid-mediated delivery of active pharmaceutical ingredients (API) opened new possibilities in advanced therapies. By encapsulating an API into a lipid nanocarrier (LNC), one can safely deliver APIs n...
doi.org
January 30, 2025 at 7:36 AM
🚀 Check out our latest review, "Computational Methods for Modeling Lipid-Mediated Active Pharmaceutical Ingredient Delivery", now published in Molecular Pharmaceutics (@ACS Journals)! 🧪📖
🔗 Read here: doi.org/10.1021/acs....
🔗 Read here: doi.org/10.1021/acs....
Reposted by Paulo C. T. Souza
So seems like the group is on a roll this month!
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
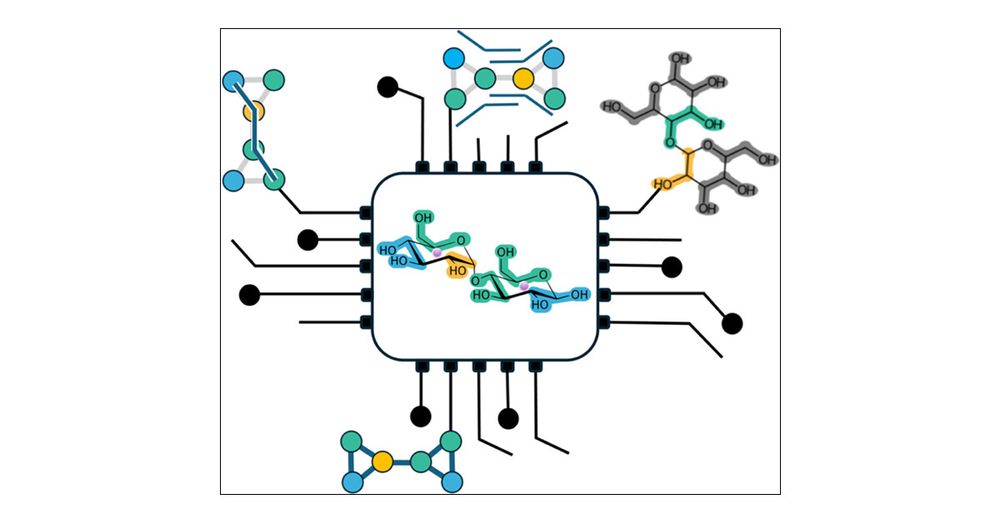
Systematic Approach to Parametrization of Disaccharides for the Martini 3 Coarse-Grained Force Field
Sugars are ubiquitous in biology; they occur in all kingdoms of life. Despite their prevalence, they have often been somewhat neglected in studies of structure–dynamics–function relationships of macro...
pubs.acs.org
January 17, 2025 at 12:27 PM
So seems like the group is on a roll this month!
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
Here’s our latest work on parametrising coarse-grained models of sugars (disaccharides) by Astrid Brandner & in collaboration with Iain Smith, @pauloctsouza.bsky.social and @cg-martini.bsky.social pubs.acs.org/doi/10.1021/... #glycotime
After some work, we’ve now made the #GitHub for our #Martini3 #LNP models and protocols publicly available: github.com/Martini-Forc...
We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation
We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation

January 3, 2025 at 9:51 AM
After some work, we’ve now made the #GitHub for our #Martini3 #LNP models and protocols publicly available: github.com/Martini-Forc...
We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation
We also have a new #ChemRxiv with some minor updates: chemrxiv.org/engage/chemr...
#LipidNanoparticles #MolecularDynamics #DrugDelivery #mRNA #Simulation
Excited to announce the release of our preprint, "The Martini 3 Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior", now available on @chemrxiv.bsky.social
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
December 26, 2024 at 1:47 PM
Excited to announce the release of our preprint, "The Martini 3 Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior", now available on @chemrxiv.bsky.social
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
Reposted by Paulo C. T. Souza
Martini 3 parameters for carbon nanomaterials, including my nanotubes.
#CompChem
chemrxiv.org/engage/chemr...
#CompChem
chemrxiv.org/engage/chemr...

Martini 3 coarse-grained models for carbon nanomaterials
The Martini model is a coarse-grained force field allowing simulations of biomolecular systems as well as a range of materials, including different types of nanomaterials of technological interest. Re...
chemrxiv.org
December 6, 2024 at 4:15 PM
Martini 3 parameters for carbon nanomaterials, including my nanotubes.
#CompChem
chemrxiv.org/engage/chemr...
#CompChem
chemrxiv.org/engage/chemr...
Excited to share that our recent research using #Martini3 #CG simulations on the role of Spc2 in the yeast signal peptidase complex (SPC) has been published in the Journal of Cell Biology.
Read the full article here: rupress.org/jcb/article/...
Read the full article here: rupress.org/jcb/article/...

November 24, 2024 at 11:01 AM
Excited to share that our recent research using #Martini3 #CG simulations on the role of Spc2 in the yeast signal peptidase complex (SPC) has been published in the Journal of Cell Biology.
Read the full article here: rupress.org/jcb/article/...
Read the full article here: rupress.org/jcb/article/...

