
Dr. Buceta at @i2sysbio.es welcome applications for 36-month postdocs on:
👉 Mechanical markers of tissue dysfunction due to ageing – digital twins & machine learning
Details here: fgcsic.es/convocatoria...
📩 Contact javier.buceta@csic.es to shape your proposal!
@epimechfc.bsky.social

Dr. Buceta at @i2sysbio.es welcome applications for 36-month postdocs on:
👉 Mechanical markers of tissue dysfunction due to ageing – digital twins & machine learning
Details here: fgcsic.es/convocatoria...
📩 Contact javier.buceta@csic.es to shape your proposal!
@epimechfc.bsky.social







🎥🌐 youtu.be/43V4MOtAf4k?...
🧪🧬🖥️ 🌾🌱🍇 #plantscience

🎥🌐 youtu.be/43V4MOtAf4k?...
🧪🧬🖥️ 🌾🌱🍇 #plantscience

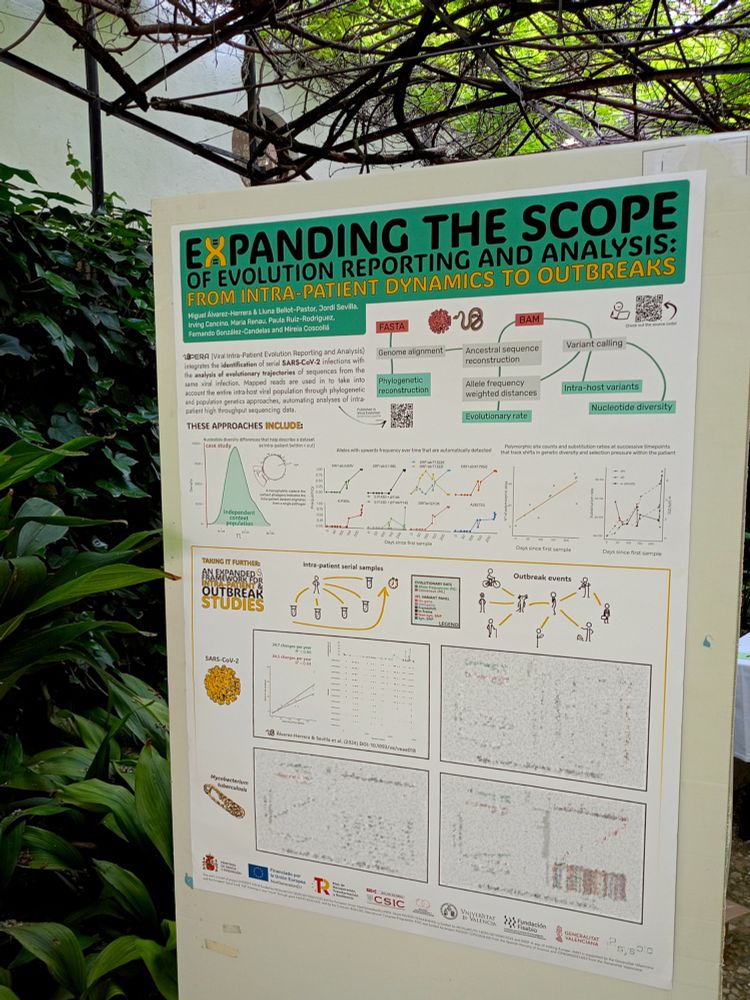
Our workflow for SARS-CoV-2 intra-patient evolution analysis now fully supports Snakemake 8/9 (with dedicated CI tests), BIONJ trees, richer reports, added config parameters, modular Snakefiles and more! 🧪👉 github.com/PathoGenOmics-Lab/VIPERA

Our workflow for SARS-CoV-2 intra-patient evolution analysis now fully supports Snakemake 8/9 (with dedicated CI tests), BIONJ trees, richer reports, added config parameters, modular Snakefiles and more! 🧪👉 github.com/PathoGenOmics-Lab/VIPERA









Desarrolla HaploScope, una pipeline híbrida (Illumina + Nanopore/PacBio) para reconstrucción de haplotipos bacterianos y virales en @i2sysbio.bsky.social
Código: JAEINT25_EX_1603
🗓️ Solicita hasta 2 mayo
🔗 rb.gy/c6c8p2
#Bioinformática #Nextflow #JAEIntro #CSIC #TFM
Desarrolla HaploScope, una pipeline híbrida (Illumina + Nanopore/PacBio) para reconstrucción de haplotipos bacterianos y virales en @i2sysbio.bsky.social
Código: JAEINT25_EX_1603
🗓️ Solicita hasta 2 mayo
🔗 rb.gy/c6c8p2
#Bioinformática #Nextflow #JAEIntro #CSIC #TFM









• get_MNV: Reclassifies SNVs as MNVs and predicts amino acid changes.
• snpick: SNP extraction for large alignments.
• VIPERA: Workflow for SARS-CoV-2 evolution.
• mycolorsTB: R color palette for M. tuberculosis.

• get_MNV: Reclassifies SNVs as MNVs and predicts amino acid changes.
• snpick: SNP extraction for large alignments.
• VIPERA: Workflow for SARS-CoV-2 evolution.
• mycolorsTB: R color palette for M. tuberculosis.
www.uv.es/institute-in...

www.uv.es/institute-in...
@i2sysbio.bsky.social available as well in bioconda bioconda.github.io/recipes/snpi... @pathogenomics.bsky.social

